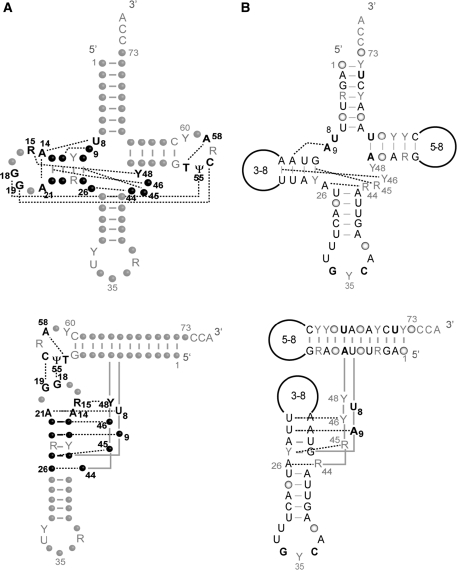
Figure 1.
Tertiary interaction networks in tRNAs. (A) Canonical tRNA (e.g. bacterial, archaeal, eukaryotic cytosolic). Data are according to crystallographic structures (5,6). Nucleotide positions involved in the nine tertiary interactions are indicated in bold. They are either strictly conserved, semi-conserved (Y for pyrimidine, R for purine) or not conserved (black dots) and bring together secondary structural domains (top) into an L-shaped 3D fold (bottom). Other strictly conserved or semi-conserved nucleotides are indicated in gray. Numbering is according to (2). Residues 8, 9 and 26 are considered as connectors between domains. (B) Mammalian mitochondrial tRNAAsp. The displayed ‘typical’ secondary and tertiary interactions have been compiled under ‘Mamit-tRNA’ data (http://mamit-trna.u-strasbg.fr/, 32) by alignment of 136 sequences. Nucleotides in black and bold characters are strictly conserved. Those indicated in black are conserved to ≥90%. Dominance of a given nucleotide in ≥50% and ≤90% of sequences as well as semi-conservation (Y or R) is indicated in grey. Conservation ≤50%, is represented by open circles. Due to large size and sequence variations in D- and T-loops, no sequence conservation can be indicated. Four tertiary interactions can be predicted in the core of the molecule. No interaction between the D- and T-loops can be anticipated in any mitochondrial tRNAAsp sequence.