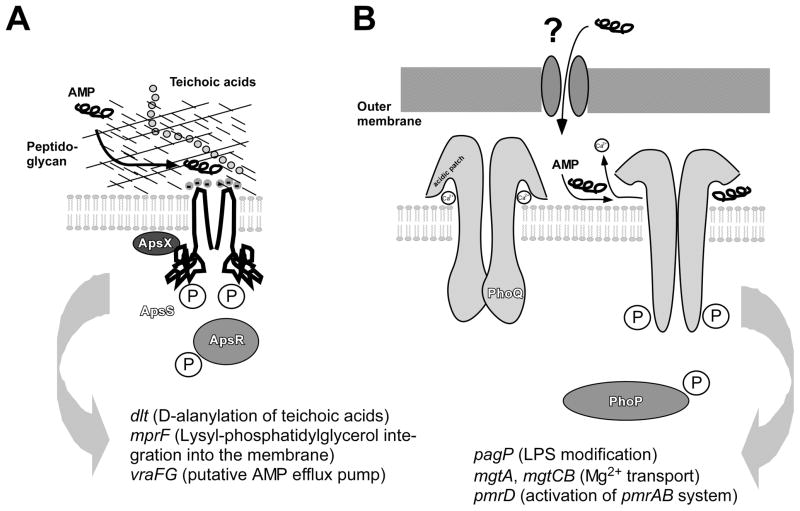
Fig. 1. The Gram-negative and Gram-positive AMP sensors.
A, The Staphylococcus aps system. The Gram-positive cell surface, containing a thick layer of peptidoglycan and anionic teichoic acids, is shown schematically. Binding of a cationic AMP leads to activation of the system in a mechanistically yet poorly understood fashion. The extracellular loop of the ApsS sensor is extremely short, carrying three negative charges that presumably are crucial for the interaction with the cationic AMP. A phosphorylation cascade as typical for two-component systems is believed to activate target gene transcription, with the ApsX protein participating in signal transduction in an unknown way. It is not known whether the ApsS sensor forms a dimer, but dimer formation of sensor proteins is common. B, The Salmonella phoP/phoQ system. Activation by CAMPs, which penetrate the Gram-negative outer membrane in a yet undefined manner, occurs by deplacement of divalent cations in an acidic patch of the PhoQ sensor. The PhoQ acidic region is much larger than the acidic extracellular loop of ApsS. This leads to structural rearrangements including notably two α-helices that move away from the membrane, and ultimately auto-phosphorylation of ApsS and subsequent phosphorylation of PhoP. Main regulatory targets of the systems are shown at the bottom. While both systems control many targets, those involved in CAMP resistance are the dlt, mprF, and vraFG loci in Staphylococcus, and the pagP locus in Salmonella. Further, the pbgPE operon, which is also involved in CAMP resistance by lipid A modification, is regulated by pmrAB, which is under control of phoPQ.