Abstract
Human metapneumovirus (hMPV) is a major cause of lower respiratory tract infections in infants, elderly and immunocompromised patients. Little is known about the response to hMPV infection of airway epithelial cells, which play a pivotal role in initiating and shaping innate and adaptive immune responses. In this study, we analyzed the transcriptional profiles of airway epithelial cells infected with hMPV using high-density oligonucleotide microarrays. Of the 47,400 transcripts and variants represented on the Affimetrix GeneChip Human Genome HG-U133 plus 2 array, 1601 genes were significantly altered following hMPV infection. Altered genes were then assigned to functional categories and mapped to signaling pathways. Many up-regulated genes are involved in the initiation of pro-inflammatory and antiviral immune responses, including chemokines, cytokines, type I interferon and interferon-inducible proteins. Other important functional classes up-regulated by hMPV infection include cellular signaling, gene transcription and apoptosis. Notably, genes associated with antioxidant and membrane transport activity, several metabolic pathways and cell proliferation were down-regulated in response to hMPV infection. Real-time PCR and Western blot assays were used to confirm the expression of genes related to several of these functional groups. The overall results of this study provides novel information on host gene expression upon infection with hMPV and also serves as a foundation for future investigations of genes and pathways involved in the pathogenesis of this important viral infection. Furthermore, it can facilitate a comparative analysis of other paramyxoviral infections to determine the transcriptional changes that are conserved versus the one that are specific to individual pathogens.
INTRODUCTION
Human metapneumovirus (hMPV) is a non-segmented negative sense RNA virus belonging to the family of Paramyxoviridae. Since its discovery in 2001, hMPV has been identified as a major cause of respiratory infections worldwide (Kahn, 2006). Virtually, all the children are infected by the age of five (van den Hoogen, de Jong et al., 2001). Around 12% of all respiratory tract infections in children are caused by hMPV (Kahn, 2006) (Williams, Harris et al., 2004) , second only to respiratory syncytial virus (RSV), another member of the Paramyxoviridae family. HMPV also accounts for 10% of all hospitalizations of elderly patients with respiratory tract infections (Falsey, Erdman et al., 2003) and is a significant pathogen in immunocompromised patients (Williams, Martino et al., 2005).
The clinical features associated with hMPV in children are similar to those of RSV. hMPV is associated with both upper and lower respiratory tract infections, as well as with asthma exacerbation (Kahn, 2003)(Kahn, 2006)(Esper, Boucher et al., 2003). Fever, cough, tachypnea, wheezing and hypoxia are frequently observed in infected children. In Children with a clinical syndrome consistent with bronchiolitis, chest radiographs demonstrate hyperinflation, as well as focal infiltrates and peribronchial cuffing (Williams, Harris et al., 2004).
As a recently identified virus, little is know about hMPV pathogenesis. Investigations done in small rodent models of infection have shown that hMPV infection induced important pulmonary inflammation, characterized by alveolitis, interstitial inflammation and increased peribronchiolitis (Alvarez, Harrod et al., 2004)(Hamelin, Yim et al., 2005)(Williams, Tollefson et al., 2005) (Wyde, Chetty et al., 2005). Airway remodeling and increased mucus production was also observed in lungs of BALB/c mice (Hamelin, Prince et al., 2006). Although hMPV shares similar epidemiological and clinical features with RSV (Principi, Bosis et al., 2006) (Williams, Wang et al., 2006), we and others have shown that they induce a different spectrum of cytokines and chemokines both in vitro, using dendritic cells, and in vivo, in a mouse model of infection, or in children, suggesting two viruses have different ability to induce cellular responses (Guerrero-Plata, Casola et al., 2005) (Jartti, van den Hoogen et al., 2002) (Laham, Israele et al., 2004).
Similar to RSV, airway epithelial cells are the primary target of hMPV infection (Biacchesi, Pham et al., 2006) (Alvarez, Harrod et al., 2004). Under normal conditions, the respiratory epithelium represents the principal cellular barrier between the environment and the internal milieu of the airways and is responsible for particulate clearance and surfactant secretion. However, after exposure to infectious agents, airway epithelial cells are able to secrete a variety of proinflammatory/immunoregulatory molecules, which induce the migration and activation of leukocytes and therefore play a key role in inflammatory and infectious processes of the lung. Here, we used high density oligonucleotide probe-based cDNA microarrays to investigate host transcriptional responses induced by hMPV infection of airway epithelial cells. For these studies, we used A549 cells, a well differentiated carcinoma-derived cell line that retains features of type II like alveolar epithelial cells (Smith, 1977). We have recently shown that hMPV-induced chemokine expression in primary small alveolar epithelial cells (SAE) and inA549 is similar, suggesting that A549 cells can be used as a model to study lower airway epithelial cell responses to hMPV infection (Bao, Liu et al., 2007). Our results show that hMPV infection induced significant changes in epithelial cell gene expression in a time-dependent fashion. Expression levels of 1601 genes from different biological groups were significantly altered over a period of 72 h. These differentially expressed genes suggested changes due to hMPV infection in signaling pathway leading to the induction of inflammatory and immunomodulatory molecules, to molecules involved in regulation of cellular signaling, gene transcription, metabolic pathways, apoptosis and antioxidant responses .The above findings will provide a foundation for future studies investigating the role of various genes and its pathways in the pathogenesis of this important respiratory virus.
RESULTS AND DISCUSSION
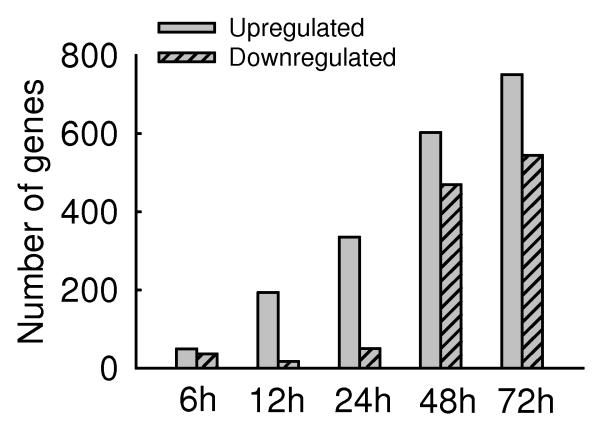
We have used high-density microarrays analysis to obtain a comprehensive profile of gene expression induced by hMPV infection. The raw data associated with gene expression profiles have been uploaded to GEO database with accession number GSE8961 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE8961). S+ Array Analyzer 2.1 a module in S-PLUS 7.0 (Insightful Inc.) was used for background correction, summarization, normalization and differential expression test. The probe level analysis was performed on the CEL files using the RMA (Robust Multichip Analysis) which performs rma background correction, quantiles normalizations and medianplish to summarize the probe sets. Probe sets absent across all the chips both control and treated were filtered resulting in 30,667 genes. One way Anova was performed on these genes and corrected for Type 1 error using Bonferroni. Gene expression was first analyzed in mock-infected cells at 12 h and 48 h p.i. We found that the transcriptional profile of these two control groups was not significantly different, indicating that gene expression did not change over time in mock-infected cells. We therefore chose the 48 h mock-infected cell to compare to infected cells at different time points p.i. Microarray analysis revealed that the expression level of 1601 genes was significantly modified (p-value ≤0.05) over a period of 72 h in infected cells, indicating that hMPV induces profound changes in global gene transcription. Fig.1A represents the number of genes that were filtered significant from One way Anova at p-value<=0.05 and whose expression changed at least two-fold in response to hMPV infection. At 6 h post-infection, a significant number of genes were already modified in response to the virus, with the number of genes up-regulated slightly higher than the number of genes down-regulated. At 12 and 24 h post-infection, up-regulated genes dominated and were nearly 13 and 5 folds more than the down-regulated genes, respectively. At 48 and 72 h time points, a total of 4.2% and 5.2% of genes were affected by hMPV infection, respectively, with a ratio of up-regulated versus down-regulated genes no less than 2.
Fig.1. Host transcriptional responses induced by hMPV.
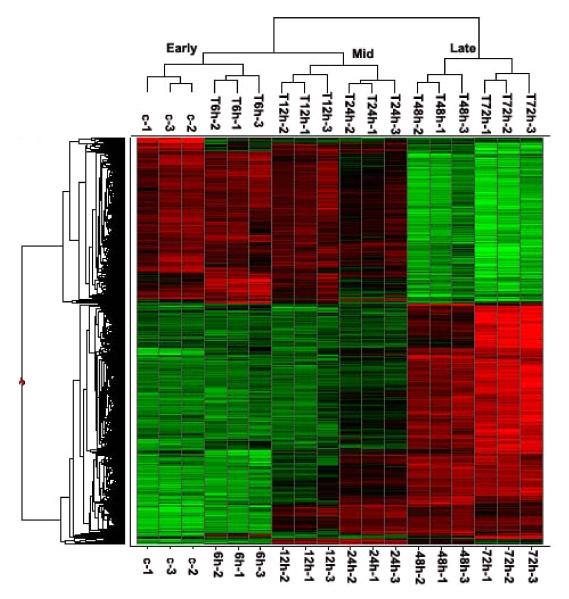
(A) Kinetics of changes in genes expression. Total RNA extracted from uninfected and hMPV-infected A549 cells were hybridized to the HG-U133 plus 2.0 GeneChip Arrays, as described in Methods. The genes, filtered as significant in response to hMPV infection at p-value<=0.05 and with a fold change of two or above, compared to baseline (uninfected), are represented at various time post-infection. Fold change is calculated on average expression values for each time point compared to the baseline. (B) Clustering and heat map analysis of hMPV-regulated genes. The expression pattern of genes significantly altered in response to hMPV infection is represented as a hierarchical clustering, using UPGMA (Unweighted Pair-Group Method with Arithmetic mean) with Euclidean distance measure. The heat map is an intensity plot which represents the clusters within the dataset. The column dendrogram represent the cluster within the time post hMPV infection and row dendrogram represent the genes clusters with similar pattern within and across time points. The red color represents the up-regulation and green the down-regulation of gene expression over time. Treatment conditions are indicated at the bottom of the figure: C represents the uninfected cells; 6 h, 12 h, 24 h, 48 h and 72 h indicate the time points of hMPV infection.
Differentially expressed genes were clustered on the expression profile over the 72 h time course of infection using Spotfire DecisionSite 9.0, (Spotfire, Somerville, MA), to help visualizing patterns of genes expression within and across clusters. The clustering algorithm in Spotfire was performed on z-score normalized expression values. Color corresponds to the expression level of the transcript with low, intermediate and high expression represented by green, black, and red, respectively (Fig.1B). The differentially expressed genes were further analyzed by K-mean clustering (n=12), to identify distinct expression patterns and correlation between functional groups within clusters, i.e., genes that perform similar functions share comparable expression profiles or their expression is regulated in a similar manner in the course of hMPV infection (supplementary Fig.1, Table1). For instance, the CXC chemokines GRO-α, GRO-β, GRO-γ, IL-8 and I-TAC clustered together in cluster 11, where genes were progressively up over the 72 h period of infection. Similarly, cluster 4 has interferon regulatory factors (IRFs), signal transducers and activators of transcription (STATs), two families of transcription factors important for the regulation of chemokines, as well as interferons (IFNs) and IFN-inducible genes together with hyperbolic pattern (peaking at 24hrs). All IFN-α genes, whose expression occur following the initial production of IFN-β, were grouped in pattern 5, where up-regulation started after 24 h and peaked at 48 h post-infection. Of interest, several antioxidant genes, i.e. microsomal glutathione S-transferase 1 (MGST1), MGST2, glutathione S-transferase A4 (GSTA4), and peroxiredoxin (PRDX)1,3 and 6 were progressively downregulated in the course of infection and clustered together in pattern 10.
Table 1.
Validation of hMPV-induced gene expression by real-time PCRa
| Gene symbol | GeneBank No. | Fold induction |
|---|---|---|
| GRO-α | NM 001511 | 9 |
| GRO-β | M57731 | 11 |
| IL-8 | NM 000584 | >20 |
| IP-10 | NM 001565 | >20 |
| RANTES | AF043341 | >20 |
| IL-6 | NM 000600 | >2 |
| IFNβ1 | NM 002176 | >20 |
| RIG-I | AI304317 | >20 |
| COX-2 | AY151286 | >20 |
| TRAF-1 | NM 005658 | 10 |
| IKBα | AI078167 | >20 |
| IKKε | NM 014002 | 5 |
| STAT-1 | BC002704 | >20 |
| IRF-1 | NM 002198 | >20 |
| NF-κB1 | M55643 | 10 |
| RELA/P65 | NM 021975 | >20 |
RNA extracted from uninfected or hMPV-infected cells for 12 h was used as template for RT-PCR, as described in Methods. Fold induction represents change in level of target gene expression, after normalization to an endogenous reference (18S) sample, in hMPV-infected versus uninfected cells.
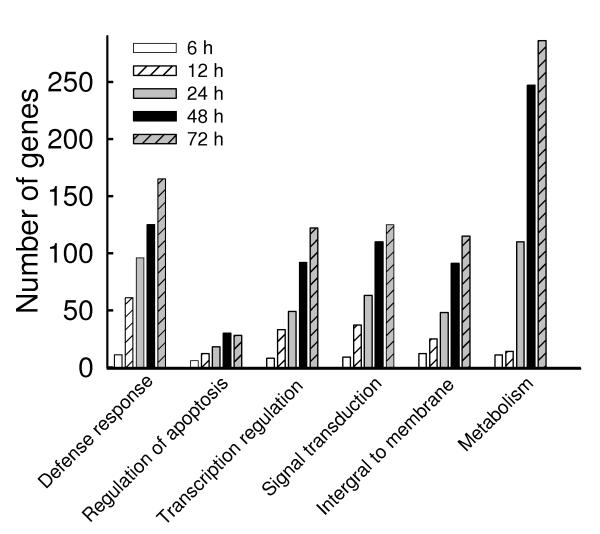
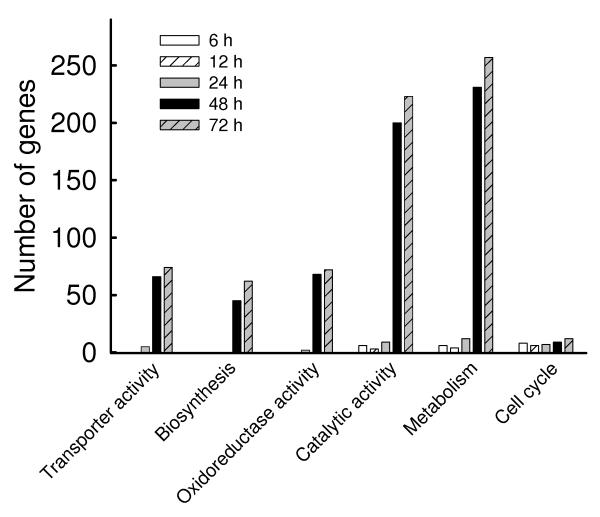
The genes with absolute fold change of 2 or more at each corresponding time point were further analyzed for functional genomics with DAVID Bioinformatics Resources (http://david.abcc.ncifcrf.gov/tools.jsp). The number of genes classified in functional groups with an enrichment score ≥1.5 and P ≤ 0.05, was plotted in Fig. 2A and Fig. 2B (Supplementary Table II), representing the up- and down-regulated genes, respectively. Overall, hMPV infection caused significant increase in the expression of genes related to defense responses, including genes necessary for innate and adaptive immune responses, regulation of gene transcription and signal transduction, while simultaneously inhibiting the expression of genes belonging to metabolic and catalytic pathways, cellular transport and cell cycle. To validate the data obtained by microarrays, we analyzed the expression of 16 different genes by real time (RT)-PCR, as shown in Table I. Induction of the chemokines RANTES, IL-8, IP-10, GRO-α and -β, the cytokines IL-6 and IFN-β, the signaling molecules RIG-I, TRAF1, IKKε, IKB-α and COX2, and the transcription factors NF-κB1/p50, RelA/p65, IRF-1 and STAT1, following hMPV infection, were all in accordance with the genes expression results. Important functional classes of differentially expressed genes in response to hMPV infection are discussed below.
Fig.2. Gene ontology analysis of hMPV-induced transcriptional responses.
The genes filtered as significant in response to hMPV infection at p-value<=0.05 and with a fold change of two or above, compared to baseline (uninfected), were analyzed using the gene ontology tool available in http://david.abcc.ncifcrf.gov and grouped using medium classification stringency. The representative groups with enrichment score of 1.5 or above and P value of 0.01 or less are presented in_(A) for the up-regulated genes and in (B) for the down-regulated genes.
Proinflammatory chemokines and cytokines
The airway epithelium forms a defense barrier against environmental pathogens and plays an important role in initialing pulmonary inflammatory and immune responses following invasion of infectious agents (Garofalo & Haeberle, 2000). Like RSV, hMPV has been shown to be a potent inducer of airway inflammation (Darniot, Petrella et al., 2005) (Hamelin, Prince et al., 2006) (Crowe, Jr. & Williams, 2003). Much of the cellular response at sites of tissue inflammation is controlled by gradients of chemotactic factors that direct leukocyte transendothelial migration and movement through the extracellular matrix. The composition of this cellular response is dependent upon the discrete target-cell selectivity of these chemotactic molecules. Chemokines are a superfamily of cytokines that regulate the migration and activation of leukocytes to the site of infections (Baggiolini, Dewald et al., 1997). They have been classified in CXC, CC, C and CX3C subfamilies based on the number and position of conserved cysteine residues (Baggiolini, 1998). Our results show that hMPV infection of airway epithelial cells induced the expression of a variety of CXC (Fig.3A) and CC (Fig.3B) chemokines, but not C or CX3C chemokines, in a time-dependent and replication-dependent manner, as UV-inactivated virus failed to up-regulate chemokine, as well as cytokine, gene expression (data not shown).
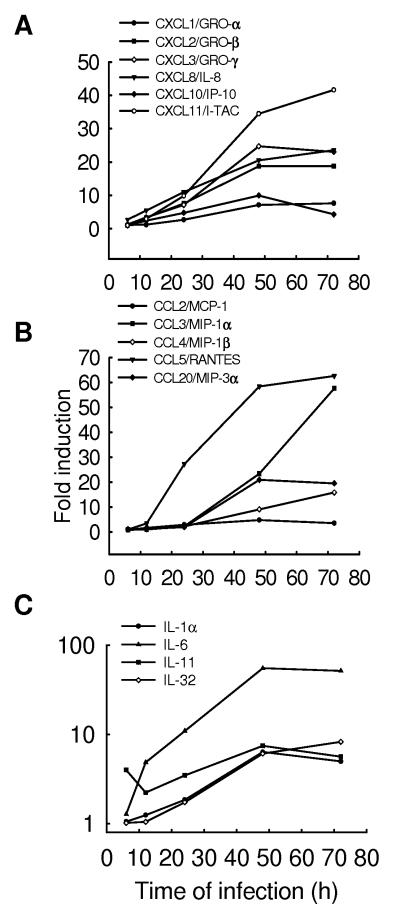
Fig.3. Changes in chemokine and cytokine expression after hMPV infection.
A549 cells were infected with hMPV at MOI of 1 for various length of time. Total RNA was extracted from control and infected cells and subjected to a microarray analysis, as described in Methods. Induction of CXC chemokines (A), CC chemokines (B) and cytokines (C) genes is plotted as a function of time. Fold change represents the ratio of gene expression level of infected versus uninfected cells at the different time points.
Similar to RSV (Zhang Y, Luxon et al., 2001), hMPV was a strong inducer of the ELR-containing CXC chemokines IL-8, GRO-α,-β and -γ, which recruit and activate neutrophils, a prominent cell type seen in early airway inflammation induced by both RSV and hMPV in experimental animal models of infection (Hamelin, Yim et al., 2005). Furthermore, significant levels of IL-8 have been detected in respiratory secretions of hMPV-infected children with bronchiolitis (Jartti, van den Hoogen, et al., 2002)(Laham, Israele et al., 2004).
Following an initial period of remarkable lung neutrophilia, mononuclear cells, including macrophages/monocytes and lymphocytes, represent the majority of the lung inflammatory cells present in animal models of hMPV infection (Hamelin, Yim et al., 2005). Non-ELR-containing CXC chemokines recruit and activate T-lymphocytes and NK cells. Among the non-ELR-containing CXC chemokines, hMPV induced the expression of IP-10 and I-TAC, which are both interferon-inducible proteins and have been shown to play a role in limiting viral replication in other infection models (Dufour, Dziejman et al., 2002)(Hamilton, Mahalingam et al., 2004). Since human airway epithelial cells constitutively express the CXCR3 receptor (Shahabuddin, Ji et al., 2006), which regulates epithelial cell movement and binds both IP-10 and I-TAC, these two chemokines may also contribute to airway repair and reconstitution.
Among the CC chemokines, hMPV induced the expression of RANTES, MCP-1, MIP-1α, MIP-1β and MIP-3α/Exodus-1. These molecules have been shown to activate a variety of cell types, including monocytes, T-lymphocytes, eosinophils, basophils and dendritic cells, and they are likely to play a significant role in the pathogenesis of hMPV-induced inflammatory lung disease, as well as in the modulation of immune responses, as it has been previously shown in RSV and other models of respiratory viral infections (Bonville, Lau et al., 2004) (Domachowske, Bonville et al., 2000)(Haeberle, Kuziel et al., 2001). In contrast to RSV (Zhang, Luxon et al., 2001), we did not detect induction of the CX3C chemokine fractalkine, a unique chemokine that fulfill the dual functions of an adhesion molecule and a chemoattractant, by recruiting NK cells, cytolytic T-cells and macrophages (Stievano, Piovan et al., 2004).
Chemokine induction in response to hMPV infection was also investigated by Bio-plex assays, shown in Table II. Similar to gene expression profiles, there was a time-dependent increase in the secretion of CXC chemokines, such as IL-8 and IP-10, and CC chemokines, such as MCP-1, MIP-1α and –β and RANTES. UV-inactivated virus stimulation did not induce a significant increase in chemokine secretion, similar to what we have previously reported (Bao, Liu et al., 2007).
Table 2.
Validation of hMPV-induced chemokines and cytokines secretion by Bio-Plex a
| Gene symbol |
hMPV 6 h |
hMPV 12 h |
hMPV 24 h |
hMPV 48 h |
hMPV 72 h |
control 12 h |
control 48 h |
UV hMPV 48 h |
|---|---|---|---|---|---|---|---|---|
| IL-8 | 104.2±8.9611 | 166.7±14.4 | 938.2±75.9 | 2293.8±106.5 | 4315.8±182.6 | 35.6±4.38 | 107.1±22.2 | 132.2±24.9 |
| IP-10 | 25.2±0.9939 | 370.7±91.1 | 1328.8±134.0 | 5333.4±109.5 | 3525.8±113.7 | 20.7±5.3 | 14.5±7.3 | 18.9±5.4 |
| MCP-1 | 583.5±95.9 | 1211.9±121.7 | 1937.2±323.1 | 2062.5±55.6 | 2167.7±156.8 | 385.8±33.1 | 1086.2±166.2 | 1186.1±145.3 |
| MIP-1α | 1.1±0.02 | 1.4±0.1 | 4.8±0.85 | 22.8±2.8 | 35.4±1.7 | 1.6±0.2 | 1.3±0.1 | 1.2±0.1 |
| MIP-1β | 3.8±0.2 | 5.0±0.3 | 6.6±0.6 | 49.8±7.4 | 61.3±6.2 | 3.6±0.2 | 3.1±0.2 | 3.0±0.1 |
| RANTES | 10.9±0.7 | 25.6±9.4 | 230.9±24.5 | 1779.3±48.3 | 1870.2±122.4 | 15.1±0.8 | 11.8±1.3 | 10.7±1.4 |
| IL-6 | 10.0±0.7 | 120.8±29.7 | 698.2±62.6 | 4061.9±43.1 | 4779.3±545.8 | 0.3±0.2 | 4.9±0.4 | 0.9±0.4 |
Supernatants from uninfected (control) cells or cells infected with live or UV-inactivated hMPV, MOI of 1, were harvested at the indicated time points of infection. Chemokine and cytokine secretion was measured by Bio-Plex assay.
Fig.3C shows hMPV-induced changes in cytokine gene expression. IL-1α and IL-6 are the prototype of pro-inflammatory cytokines, whose inducible secretion has been recently demonstrated both in animal models and in children with bronchiolitis in response to hMPV infection (Guerrero-Plata, Casola, & Garofalo, 2005)(Laham, Israele et al., 2004). Similar to RSV, hMPV infection has been associated with wheezing and also with airway hyperresponsivenes, in a mouse model of infection (Hamelin, Prince et al., 2006). IL-11 is a cytokine that has been linked to airway remodeling, as well as airway hyperresponsiveness (Zheng, Zhu et al., 2001). It has been detected in vivo in children with detectable wheezing and viral infections, including RSV (Einarsson, Geba et al., 1996), suggesting that IL-11 can play an important role in the pathogenesis of hMPV-induced lung disease. IL-32 is a recently identified cytokine, which can be considered pro-inflammatory as it induces expression of multiple cytokines and chemokines in stimulated cells (Kim, Han et al., 2005). This is the first time that a viral infection has been shown to induce IL-32 expression in airway epithelial cells, although it can be produced by epithelial cells in response to interferon stimulation (Kim, Han et al., 2005).
Interferon response
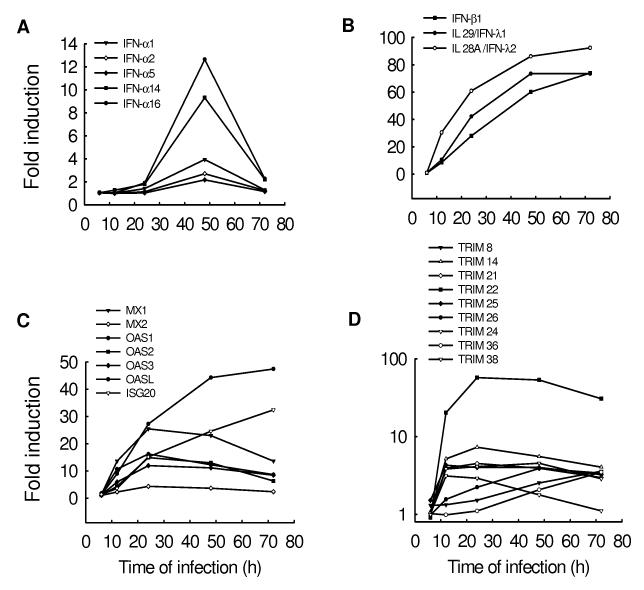
Interferons (IFNs) are a superfamily of cytokines with antiviral, as well as antiproliferative and immunomodulatory functions (Smith, Lombardi et al., 2005). Type I IFNs, which include IFN-α,-β and –λ, are the first line of defense against viral infections and their expression is tightly regulated [Reviewed in (Stetson & Medzhitov, 2006)].. Their activity depend on the expression of a large set of interferon-stimulated genes (ISGs), of which Mx proteins, 2-5 oligoadenylate synthetase (OAS), and double-stranded RNA-activated protein kinase R (PKR) are the best characterized (Smith, Lombardi, & Foster, 2005). Microarray analysis showed that hMPV infection induced very significant changes in type I IFN and ISG gene transcription (Fig.4). IFN-β, -λ1 and –λ2 (also known as IL-28 and -29) genes were the first to be induced, starting between 6 and 12 h post-infection, with a continuous increase in their level of expression up to 72 h (Fig.4B). Induction of IFN-α genes was lower and more transient, compared to IFN-β and –λ, starting at 24 h, peaking at 48 h, and returning almost back to control levels by 72 h post-infection (Fig.4A). This pattern of expression is in agreement with the knowledge that IFN-β gene expression is a primary cellular response to viral infection and with the recent findings that IFN-β and –λ genes share a common mechanism of induction (Onoguchi, Yoneyama et al., 2007), which is dependent on activation of the transcription factors NF-κB, IRF-3 and AP-1 (Hiscott, Pitha et al., 1999), constitutively expressed in infected cells. On the other hand, IFN-α gene expression is a secondary event in the course of the infection and depends on IFN-β induction of the transcription factor IRF-7 (Sato, Suemori et al., 2000). These results confirms our previous findings in vitro and in vivo that hMPV is a better inducer of IFN-α than RSV, which does not upregulate IFN-α expression in either epithelial cells (Spann, Tran et al., 2004) or in monocyte-derived dendritic cells (Guerrero-Plata, Casola et al., 2006) and induce lower levels of IFN-α in a mouse model of infection (Guerrero-Plata, Baron et al., 2005), likely due to the absence in the hMPV genome of the two non-structural (NS) proteins NS1 and NS2, which have been reported to inhibit type I IFN-mediated signaling (Ramaswamy, Shi et al., 2006) (Lo, Brazas et al., 2005)(Spann, Tran et al., 2005).
Fig.4. Changes in the expression of interferons (IFN) and interferon stimulated genes (ISGs) after hMPV infection.
A549 cells were infected with hMPV at MOI of 1 for various length of time. Total RNA was extracted from control and infected cells and subjected to a microarray analysis, as described in Methods. Induction of IFN-α (A), IFN-β and -λ (B), ISG (C) and TRIM (D) genes is plotted as a function of time. Fold change represents the ratio of gene expression level of infected versus uninfected cells at the different time points.
hMPV-induced expression of type I IFNs was paralleled by the upregulation of several important antiviral genes, including Mx proteins, OASs and ISG20, as shown in Fig.4C. Mx proteins are a family of dynamin-like GTPases that mediate their effect by sequestering viral nucleocapsids, making them not accessible for viral replication. An antiviral effect of MxA has been demonstrated for various types of viral infections, mostly due to negative sense RNA viruses, including orthomyxoviruses and paramyxoviruses, while the anti-viral function of MxB has not been well defined [Reviewed in (Nagata & Mibayashi, 1997)]. MxA protein is specifically induced by type I IFNs and is a reliable biomarker of IFN-β activity (Pachner, Bertolotto et al., 2003). Promoter sequence analysis indicated that MxA contains two functional IFN-stimulated response elements (ISRE), common to many ISG gene promoters, which are essential for IFN-induced gene transcription (Ronni, Matikainen et al., 1998). MxA gene was significantly up-regulated (25 folds) in response to hMPV infection, while MxB gene was induced at a much lower level, suggesting that MxA is the protein likely to play a role as an antiviral ISG in response to hMPV.
2′-5′-oligoadenylate synthetases (OASs) were among the first interferon-induced antiviral enzymes to be discovered. Activation of OAS has been associated with many viral infections, including West Nile virus, influenza A and RSV (Ronni, Matikainen et al., 1997) (Torrence, 1999) (Lucas, Mashimo et al., 2003). The function of 2′-5′ oligoadenylate synthetase is to produce 5′-phosphorylated, 2′-5′-linked oligoadenylates (2-5A) and activate RNase L, which is required for IFN-mediated antiviral response (Malathi, Paranjape et al., 2005). hMPV infection induced the expression of all four members of the OAS family, in particular of the OASL protein, a recently identified protein whose function is still relatively unknown (Eskildsen, Hartmann et al., 2002).
Similarly, hMPV was a strong inducer of ISG20, a new 3′-->5′ exoribonuclease with activity specific for single-stranded RNA, that represents a novel antiviral pathway against viral infections (Espert, Degols et al., 2003). Its cellular expression confers resistance to a variety of RNA viruses, including vesicular stomatitis virus (VSV), influenza virus, and encephalomyocarditis virus, but not to DNA viruses (Espert, Degols et al., 2003). Whether ISG20 has antiviral activity in the contest of hMPV infection needs further investigation.
Among the interferon inducible genes, hMPV induced the expression of several members of the tripartite motif (TRIM) family, which consists of proteins characterized by a tripartite structure (Fig.4D). Several of these proteins have been identified as mediators of antiviral responses to retroviruses, in particular HIV [Reviewed in (Nisole, Stoye et al., 2005)]. However, since TRIM proteins can form high order molecular weight structures located in various cellular compartments, they have the capability of interfere with a variety of viral infections, representing a new class of antiviral proteins involved in innate immune responses. Among the TRIM proteins induced by hMPV infection, TRIM 22, also known as Staf50, showed the highest level of expression, similar to IFN-β, -λ1 and –λ2 genes. It is intriguing to speculate that TRIM22 could play a role in regulating hMPV antiviral cellular state. To investigate whether the expression of interferon and interferon-dependent genes would affect hMPV replication, we determined net production of viral infectious particles at various time points p.i. Cells were infected at MOI of 1, cell supernatant was collected every 24 h and totally replaced each time with fresh media, up to 72 h p.i., to measure viral titers. There was a progressive decrease in total viral particles production, from 24 to 72 h p.i., suggesting that viral replication is inhibited during the time course of infection (5.1±0.7×103 at 24 h, 6.2±0.5×102 at 48 h and 7.2±0.7×101 pfu/ml at 72 h p.i.). We are currently investigating this possibility using different approaches including overexpression studies and gene expression silencing by siRNA.
Transcription factors
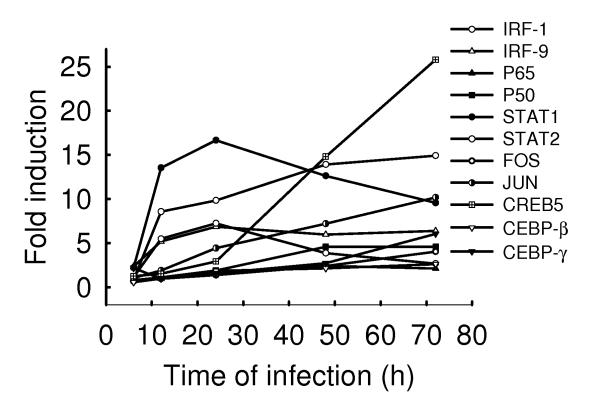
We have previously shown that RSV infection of airway epithelial cells activates several families of transcription factors involved in the regulation of proinflammatory and immunomodulatory mediators (Casola, Garofalo et al., 2001) (Casola, Garofalo et al., 2000) (Liu, Castro et al., 2003). Microarray analysis shows that hMPV induced the expression of many transcription factors, including nuclear factor (NF)-κB, interferon regulatory factors (IRFs), activating protein (AP)-1, cyclic AMP responsive element-binding (CREB) proteins, C/EBPs and signal transducers and activators of transcription (STATs) (supplementary table II and Fig.5). NF-κB is a well-characterized family of proteins involved in the expression of chemokines (e.g. IL-8 and RANTES), adhesion molecules (e. g., ICAM, VCAM, E-selectin), inducible enzymes (COX-2 and iNOS), growth factors, acute phase proteins, and receptors [Reviewed in (Nam, 2006)]. Among the different NF-κB family members, hMPV induced the expression of the p50 and p65 (also known as RelA) subunits, which we have previously shown to play a critical role in modulation of cellular and inflammatory/immune processes in response to RSV infection (Haeberle, Casola et al., 2004) (Tian B, Zhang Y et al., 2002). We have recently shown that hMPV infection of A549 cells induces nuclear translocation and DNA-binding of both p50 and p65 NF-κB subunits, indicating that both proteins are activated in response to the infection (Bao, Liu et al., 2007). Their specific role in hMPV infection is currently being investigated.
Fig.5. Induction of transcription factors after hMPV infection.
A549 cells were infected with hMPV at MOI of 1 for various length of time. Total RNA was extracted from control and infected cells and subjected to a microarray analysis, as described in Methods. Induction of transcription factor genes is plotted as a function of time. Fold change represents the ratio of gene expression level of infected versus uninfected cells at the different time points.
IRFs regulate the expression of type I IFN and chemokines [Reviewed in (Barnes, Lubyova et al., 2002)]. IRF-1 and -3 are necessary for RSV-induced RANTES gene transcription (Casola, Garofalo et al., 2001), and they also bind to the ISRE of the IFN-β gene promoter (Hiscott, Pitha et al., 1999). Binding of IFN-β to its receptor activates STAT1 and STAT2, which, together with IRF-9, form the ISGF3 complex, responsible for the expression of a variety of ISGs, including IRF-7, which is required for IFN-α gene expression (Sato, Suemori et al., 2000). HMPV-induced IRF-1 expression fell in the same cluster of IFN-β and –λ, with significant induction starting at 12 h post-infection, suggesting that regulation of these two IFN genes are IRF dependent (data not shown). Similarly, hMPV-induced STAT1 and STAT2 and IRF-9 cluster with several of the interferon-induced genes, including OAS, MxA and TAP, which are part of pattern 4 gene expression, shown in supplementary Fig.1. We have recently shown that hMPV infection induces activation of IRF-1, 3, 7 and 9, as well as of STAT1 and 2 (Bao, Liu et al., 2007), and we are currently investigating their role in hMPV-induced signaling.
Other than NF-κB, IRFs and STATs, hMPV infection induced the expression of AP-1-binding transcription factors (Jun and FOS), CREB proteins (ATF3 and CREB 5), as well as C/EBP proteins (C/EBPβ, also known as NF-IL6 and C/EBPγ), all of which have been previously shown to play an important role in RSV-induced cytokine and chemokine gene expression (Casola, Garofalo et al., 2001)(Casola, Garofalo et al., 2000)(Jamaluddin, Garofalo et al., 1996). Further studies will define their contribution to hMPV-induced gene expression.
Signal transduction
Detection of viral infection and subsequent activation of intracellular signaling pathways is virus- and cell type-dependent. Toll-like receptors (TLRs) and cytoplasmic RNA binding proteins, including IFN-induced double-stranded RNA-dependent protein kinase (PKR) and RNA helicases, such as retinoic acid-inducible gene-I (RIG-I) and melanoma differentiation associated gene-5 (MDA-5), have been identified as the major molecules responsible for recognition of viral infection and activation of signaling cascades (Aderem & Ulevitch, 2000) (Akira, Uematsu et al., 2006) (Akira & Takeda, 2004) (Kawai & Akira, 2006a) (Takeda & Akira, 2005). TLRs have been shown to activate innate immune response by recognizing different pathogen-associated molecular patterns (PAMPs). So far, 10 members of TLRs have been identified in humans, and 13 in mice. Among those, TLR3, 4, 7, 8 and 9 have been shown to be involved in the innate response to viral infections [Reviewed in (Kawai & Akira, 2006b) (Takeda & Akira, 2005)]. TLR signaling is initiated by the interaction between the cytoplasmic domain of TLR with TIR-domain-containing cytosolic adaptors. Myeloid differentiation primary response protein 88 (MyD88) is a common TIR-domain-containing adaptor of all TLRs except for TLR-3 (Pandey & Agrawal, 2006) (Kaisho & Akira, 2006) (Akira, Uematsu, & Takeuchi, 2006) (Yamamoto, Sato et al., 2003a) (Kawai & Akira, 2006a). During viral infection, MyD88 recruits member of the interleukin-1 receptor-associated kinase-1 and/or - 4 (IRAK), to activate transcription factors belonging to the NF-κB and AP-1 family via TRAF6 (Yang, Puel et al., 2005) (Jefferies, Bowie et al., 2001) (Sato, Takahashi et al., 2004). TLR-3 and -4 mediated activation of NF-κB and IRFs, in part through induction of the IκB kinase (IKK)-like molecules IKKε/TBK-1, occurs through TRIF and TRAM adaptor molecules, respectively (Kawai & Akira, 2006b) (Yamamoto, Sato et al., 2003b).
Among the non-TLR cellular viral sensors, the RNA helicases RIG-I and MDA-5 have been shown critical to detect viral infections and trigger cellular signaling in a TLR-independent manner (Kato, Sato et al., 2005)(Kato, Takeuchi et al., 2006) (Yoneyama, Kikuchi et al., 2004). Both RIG-I and MDA-5 share an homologous CARD domain (Hiscott, Lin et al., 2006) (Johnson & Gale, Jr., 2006), important for interaction with the recently identified mitochondrial adaptor protein MAVS (also known as IPS-1 or VISA/Cardif), which mediates IRF and NF-κB activation, in part through IKKε/TBK-1 induction (Hiscott, Lin, Nakhaei, & Paz, 2006) (Kawai, Takahashi et al., 2005).
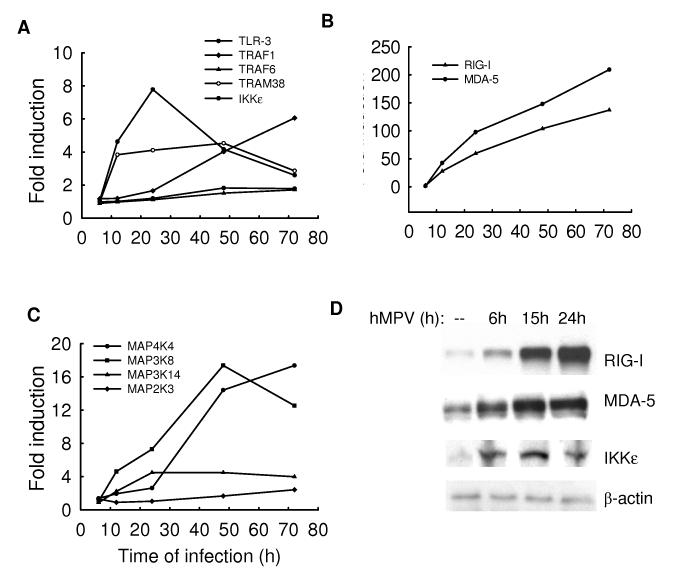
Microarray analysis shows that hMPV infection induced the expression of TLR-3, some of the TLR-adaptor molecules, RIG-I and MDA-5, as well as downstream signaling molecules such as IKKε (supplementary Table II and Fig.6A and B), suggesting that hMPV might use both TLR-dependent and -independent pathways to activate signaling cascades in airway epithelial cells, similar to what we have recently shown for RSV infection (Liu, Jamaluddin et al., 2007). Western blot analysis confirmed the expression of RIG-I, MDA5 and IKKε (Fig.6D) and preliminary studies indeed suggest that both RNA helicases and their downstream effector molecules MAVS and IKKε play a significant role in hMPV-induced gene expression (S. Liao, personal communication). The role of TLR s in hMPV-dependent cellular signaling is currently being investigated.
Fig.6. Induction of TLRs, RNA helicases and MAPK signaling cascade molecules after hMPV infection.
A549 cells were infected with hMPV at MOI of 1 for various length of time. Total RNA was extracted from control and infected cells and subjected to a microarray analysis, as described in Methods. Induction of TLR-related signaling molecules (A), RNA helicases (B), and signaling molecules related to the MAPK signaling cascade (C) is plotted as a function of time. Fold change represents the ratio of gene expression level of infected versus uninfected cells at the different time points. (D) Total cell lysates, prepared from A549 cells uninfected or infected with hMPV, MOI of 1, RSV for 6, 12 and 24 h were resolved on 10% SDS-PAGE and Western blot was performed using antibodies against RIG-I, MDA-5 and IKKε. Membranes were stripped and reprobed for β-actin as an internal control for protein integrity and loading.
Mitogen-activated protein kinase (MAPK) cascade represents a multifunctional signaling network that regulates a variety of cellular functions, in response to pathogen exposure, including the expression of cytokines and chemokines, cell growth, differentiation, and apoptosis (Seger & Krebs, 1995) (Fenton & Sinclair, 1999). The characteristic feature of this pathway is that it is initiated by a serine/threonine kinase called MAP kinase kinase kinase (MAPKKK), which then activates a unique threonine-tyrosine kinase, MAP kinase kinase (MAPKK). The latter activates the molecules of the MAP kinase family, of which extracellular signal-regulated kinase (ERK), p38 and c-Jun-terminal kinase (JNK) are the best characterized members (Karin, 1998b). They regulate activation of transcription factors belonging to the AP-1, CREB and NF-κB families (Karin, 1998a)(Karin, 1995) (Karin & Delhase, 1998). From the microarray analysis, we identified several members of MAPK pathway that respond to hMPV infection, including MAP kinase kinase 3 (MAP2K3), kinase kinase kinase 8 and 14 (MAP3K8 and 14) and kinase kinase kinase kinase 4 (MAP4K4) (Fig.6C). MAPK cascade activation plays a critical role in cellular responses to viral infections, including influenza A, varicella zoster virus and herpes virus (Zachos, Clements et al., 1999) (Ludwig, Ehrhardt et al., 2001) (Zapata, Nakatsugawa et al., 2006). We and other have shown that RSV induces activation of ERK and p38, which both play a role in RSV-induced chemokine gene expression (Chen, Monick et al., 2000) (Pazdrak, Olszewska-Pazdrak et al., 2002). Whether MAPK pathway is involved in hMPV-induced cellular responses remains to be clarified.
Antioxidants
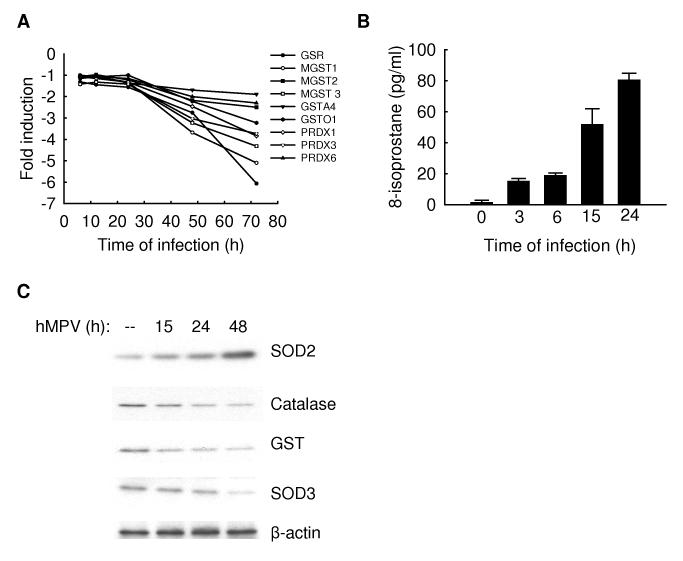
Reactive oxygen species (ROS) are ubiquitous, highly diffusable and reactive molecules produced as a result of reduction of molecular oxygen, including species such as hydrogen peroxide, superoxide anion, and hydroxyl radical, and they have been implicated in damaging cellular components like lipids, proteins and DNA. Cells are protected against oxidative damage by well developed enzymatic and nonenzymatic antioxidant systems, including superoxide dismutase (SOD), catalase, glutathione-dependent enzymes, thioredoxin, and peroxiredoxins, which protect cells against ROS and cytotoxic products of lipid peroxidation. Inducible ROS generation has been shown following stimulation with a variety of molecules, like cytokines and growth factors, and infection with certain viruses, like HIV, Hepatitis B, influenza, rhinovirus and RSV [reviewed by (Schwarz, 1996)] Indeed, oxidative stress has been shown to play an important role in the pathogenesis of acute and chronic lung diseases, including respiratory infections, as we have recently demonstrated for RSV(Castro, Guerrero-Plata et al., 2006) (Indukuri, Castro et al., 2006). Among the functional groups downregulated by hMPV infection, we found that genes involved in cellular antioxidant responses were significantly overrepresented (Supplementary Table II and Fig.7A), suggesting that hMPV infection could lead to cellular oxidative damage. To determine whether hMPV induced oxidative stress in airway epithelial cells, we investigated F2-isoprostane production in A549 cells either uninfected or infected with hMPV at various time points post-infection (p.i.). There was a progressive increase in F2-isoprostane levels in hMPV-infected cells at all time points, with a ~ 20 and 30 fold increase at 15 and 24 h p.i. respectively, when compared to control cells (Fig.7B). Similarly, there was a significant increase in other lipid peroxidation products, such as malondialedyde (MDA) or 4-hydroxynenal (4-HNE), at various time points of infection (data not shown), indicating that hMPV induces significant oxidative stress in airway epithelial cells.
Fig.7. Downregulation of antioxidant gene expression after hMPV infection.
(A). Total RNA from control or infected A549 cells was subjected to a microarray analysis. The reduced induction of antioxidant genes is plotted as a function of time. Fold change represents the ratio of gene expression level of infected versus uninfected cells at the different time points. (B). A549 cells were infected with hMPV, MOI of 3, and harvested at 3, 6, 15 and 24 h post-infection to measure F2–isoprostanes. The figure is representative of two different experiments done in triplicate. (C). Total cell lysates, prepared from A549 cells uninfected or infected with hMPV, MOI of 3, for 15,24 and 48 h were resolved on 10% SDS-PAGE and Western blot was performed using antibodies against SOD 2, 3, catalase and GST. Membranes were stripped and reprobed for β-actin as an internal control for protein integrity and loading.
To investigate the effect of hMPV infection on the expression of AOE in A549 cells, as a protective mechanism against oxidative damage, superoxide dismutase (SOD 1, 2 and 3), catalase and glutathione S-transferase (GST) protein expression was evaluated by Western blot analysis. A549 cells were infected with hMPV, MOI of 1, and harvested at 15, 24 and 48 h p.i. There was a progressive decrease of SOD 3, catalase and GST expression levels in infected A549 cells, compared to uninfected cells, while SOD 2 increased with the progression of viral infection (Fig.7C). There was no change in SOD 1 expression (data not shown). These data strongly suggest that the cellular antioxidant capacity is diminished in response to hMPV infection leading to significant oxidative stress in airway epithelial cells.
In summary, microarray studies can yield considerable novel information on the host cellular responses to hMPV infection and can serve as a foundation for future investigations aimed to understand the role of the identified genes and pathways in initiating antiviral, as well as innate and adaptive immune responses to this important pathogen.
MATERIALS AND METHODS
Cell culture and viral preparation
LLC-MK2 cells were maintained in MEM supplemented with 10% Fetal Bovine Serum (FBS), Penicillin and Streptomycin (100U/ml). A549 cells were maintained in F12K medium containing 10% (v/v) FBS, 10 mM glutamine, 100 IU/ml penicillin and 100 μg/ml streptomycin. Cell cultures were routinely screened for mycoplasma contamination. Viral stocks were obtained by infecting LLC-MK2 cells at MOI of 0.1. Around day 5-7 post-infection, cells were disrupted using glass beads, and then scraped completely from the flasks. Total medium was collected and sonicated on ice for 5 minutes. The preparation was then vortexed and centrifuged at 2900g for 15 minutes on ice. Crude virus was collected and then purified on a sucrose cushion. Viral titer was determined by immunostaining, as previously described (Guerrero-Plata, Casola et al., 2006). Viral preparations were routinely tested for LPS and cytokine contamination. Confluent cells were infected with hMPV in serum-free media with 1.0 μg trypsin/ml at a multiplicity of infection (MOI) of 1, which leads to 65-70% infected cells at 48 h p.i.. Mock-infected cells, defined as control or uninfected cells throughout the manuscript, were treated with same amount of sucrose and the same viral infection media.
High-density oligonucleotide probe-based microarrays
Confluent monolayers of A549 cells were infected with hMPV at MOI of 1 in serum-free media and harvested at 6, 12, 24, 48, or 72 hours post-infection to extract total RNA using RNAqueous®-Midi Kit (Ambion, Austin, TX), according to manufacturer’s instruction. Ten microgram of total RNA from control or infected cells was first converted to cDNA utilizing a T7-oligo (dT) primer. The synthesized cDNA was then used as a template to synthesis biotinylated cRNA, using bacteriophage T7 RNA polymerase. The biotinylated cRNA was fragmented to a mean size of 200 bases and hybridized to HG-U133 plus 2.0 Gene Chip array (Affymetrix, Santa Clara,CA), containing 47,400 sequenced human genes. Hybridization of HG-U133 plus 2.0 Array was performed at 45 °C for 16 h. Staining with phycoerythrin-streptavidin was then performed after non-stringent and stringent washes. Gene Chip arrays were scanned using a Gene Array Scanner (Hewlett Packard). All cRNA fragmentation, hybridization and scanning steps were performed at the Genomics Core of Sealy Center Molecular Science, University of Texas Medical Branch, Galveston, Texas, as previously described (Tian, Zhang et al., 2002) (Zhang, Luxon et al., 2001) Experiments were repeated in triplicate to investigate the reproducibility of gene expression profiling.
Microarray Data Processing
S+ Array Analyzer2.1 (S+AA) a statistical package from S-PLUS7.0 (Insightful Corporation, Seattle, WA) was used to analyze the microarray data. This software has the enhanced data import, QC diagnostics, normalization and differential testing methods for array analysis. Probe level data analysis was performed with CEL files. The probe level data was first subjected to pre-processing (correcting for background correction and non-specific binding, summarization) and normalization steps. The Robust Multichip Analysis (RMA) method in S+ Array Analyzer returns data that has been corrected using rma background correction, PM correction, quantiles normalization and median polish to summarize the probe sets. The quantiles method for normalization assumes an underlying common distribution, thus normalizing the chips so that their quantiles have the same value. One way Anova (Analysis of Variance) was performed on the data processed through RMA method (Irizarry, Hobbs et al., 2003). ANOVA was done on various time points of hMPV infection (6, 12, 24, 48 and 72 hrs) and using mock infection at 48 h as baseline. Bonferroni (Family-Wise Error Rate) was used to control for Type 1 errors. Data were further analyzed with clustering algorithm (Spotfire Decision Site9.0, Spotfire Inc, Somerville, MA) to study the patterns of gene expression induced by HMPV infection. Microarray raw data and analysis have been deposited in the NCBI GEO database under the accession number GSE8961.
Functional Analysis
To map the genes from gene lists to GENE Ontology categories and calculate whether these categories are over-represented in the gene lists compared to their occurrence on the chip, functional annotation clustering tool from DAVID Bioinformaticx Resources 2007, NIAID/NIH (http://david.abcc.ncifcrf.gov/summary.jsp) was used (http://scholar.google.com/scholar?num=100&hl=en&lr=&cites=17546728891026140932). The list of differentially expressed genes, up- or down-regulated by ≥2 fold in hMPV-infected cells, was uploaded into the tool. The categories with enrichment score ≥1.5 and P ≤0.05 were organized into a table (Supplementary Fig.2).
Real-time PCR
Total RNA from cells, uninfected or infected, was prepared as described above. For amplification of specific gene mRNA, Quantitative Real Time PCR (Q-RT-PCR) Applied Biosystems assays-on-demand 20× mix of primers and TaqMan MGB probes (FAM-dye labeled) for target genes and 18S rRNA (VIC-dye labeled probe) TaqMan assay reagent (P/N 4319413E) for controls were used. Separate tubes (singleplex) one-step RT-PCR was performed with 80 ng RNA for both target genes and endogenous control. The cycling parameters for one-step RT-PCR were: reverse transcription at 48 °C for 30 min, AmpliTaq activation at 95 °C for 10 min, denaturation at 95 °C for 15 s and annealing/extension at 60 °C for 1 min (repeat 30 times) on ABI7000. Duplicate CT values were analyzed in Microsoft Excel using the comparative CT (ΔΔCT) method as described by the manufacturer (Applied Biosystems, Foster City, CA). The amount of target (2–ΔΔCT) was obtained by normalizing to endogenous reference (18S) sample.
Bio-Plex
Chemokines and cytokines (IL-1RA, IL-1β, IL-2, IL-4, IL-5, IL-6, IL-7, IL-9, IL-10, IL-12 p70, IL-13, IL-17, G-CSF, GM-CSF, IFN-γ, IP-10 ,EOTAXIN, MIP-1α, MIP-1β, GCSF, FGFB, PDGF, VEGF and TNF-α) were quantified by the Luminex-based Bio-Plex system (Bio-Rad Laboratories, Hercules, CA) according to the manufacturer’s instructions. The lower limit of detection for all cytokines measured by Bio-Plex is 3 pg/ml.
Western blot analysis
Total cell lysates from uninfected and infected A549 cells were prepared using RIPA buffer, as previously described (Garofalo, Sabry et al., 1996). Proteins were normalized by protein assay (Bio-Rad, Hercules, CA), fractionated by SDS-PAGE, and transferred to polyvinylidene difluoride membranes. Membranes were blocked with 5% milk in TBS-Tween and incubated proper primary antibodies according to manufacturer’s instruction. The primary antibodies for RIG-I and MAVS were a generous gift from Dr Julkunen, National Public Health Institute, Finland. The antibody against MDA-5 was from Imgenex (San Diego, CA). Antibodies for superoxide dismutase (SOD) 1, 2 3 and catalase were from Stressgen Bioreagents (Victoria, BC, Canada). Antibody for glutathione-s-transferase (GST) was a generous gift of Dr Amasthi, Biochemistry & Molecular Biology, UTMB. Appropriate peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology) were used after primary antibody incubation. Proteins were detected by autoradiography using ECL regular or plus (Amersham Pharmacia Biotech) according to manufacturer‘s protocol. Equal loading of proteins was evaluated by stripping and reprobing the membranes with β-actin antibody.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants NIEHS 06676 and NIAID P01 062885, a pilot project from the Sealy Center for Vaccine Development at UTMB and the VRPRU N01 AI30039. X. B. was supported by the NIAID training grant T32 AI07536.
Reference List
- Aderem A, Ulevitch RJ. Toll-like receptors in the induction of the innate immune response. Nature (London) 2000;406:782–787. doi: 10.1038/35021228. [DOI] [PubMed] [Google Scholar]
- Akira S, Takeda K. Functions of toll-like receptors: lessons from KO mice. C.R.Biol. 2004;327:581–589. doi: 10.1016/j.crvi.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell (Cambridge MA) 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Alvarez R, Harrod KS, Shieh WJ, Zaki S, Tripp RA. Human metapneumovirus persists in BALB/c mice despite the presence of neutralizing antibodies. J.Virol. 2004;78:14003–14011. doi: 10.1128/JVI.78.24.14003-14011.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggiolini M. Chemokines and leukocyte traffic. Nature (London) 1998;392:565–568. doi: 10.1038/33340. [DOI] [PubMed] [Google Scholar]
- Baggiolini M, Dewald B, Moser B. Human chemokines: an update. Annual Review Immunology. 1997;15:675–705. doi: 10.1146/annurev.immunol.15.1.675. [DOI] [PubMed] [Google Scholar]
- Bao X, Liu T, Spetch L, Kolli D, Garofalo RP, Casola A. Airway epithelial cell response to human metapneumovirus infection. Virology. 2007;368(1):91–101. doi: 10.1016/j.virol.2007.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes B, Lubyova B, Pitha PM. On the role of IRF in host defense. J Interferon Cytokine Res. 2002;22:59–71. doi: 10.1089/107999002753452665. [DOI] [PubMed] [Google Scholar]
- Biacchesi S, Pham QN, Skiadopoulos MH, Murphy BR, Collins PL, Buchholz UJ. Modification of the trypsin-dependent cleavage activation site of the human metapneumovirus fusion protein to be trypsin independent does not increase replication or spread in rodents or nonhuman primates. J.Virol. 2006;80:5798–5806. doi: 10.1128/JVI.00294-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonville CA, Lau VK, DeLeon JM, Gao JL, Easton AJ, Rosenberg HF, Domachowske JB. Functional antagonism of chemokine receptor CCR1 reduces mortality in acute pneumovirus infection in vivo. J.Virol. 2004;78:7984–7989. doi: 10.1128/JVI.78.15.7984-7989.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik S, Peters B, Ebeling C, Holzhutter H. Cytosolic processing of proteasomal cleavage products can enhance the presentation efficiency of MHC-1 epitopes. Genome Inform. 2004;15:24–34. [PubMed] [Google Scholar]
- Casola A, Garofalo RP, Haeberle H, Elliott TF, Lin A, Jamaluddin M, Brasier AR. Multiple cis regulatory elements control RANTES promoter activity in alveolar epithelial cells infected with respiratory syncytial virus. Journal of Virology. 2001;75:6428–6439. doi: 10.1128/JVI.75.14.6428-6439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casola A, Garofalo RP, Jamaluddin M, Vlahopoulos S, Brasier AR. Requirement of a novel upstream response element in RSV induction of interleukin-8 gene expression: stimulus-specific differences with cytokine activation. Journal of Immunology. 2000;164:5944–5951. doi: 10.4049/jimmunol.164.11.5944. [DOI] [PubMed] [Google Scholar]
- Castro SM, Guerrero-Plata A, Suarez-Real G, Adegboyega PA, Colasurdo GN, Khan AM, Garofalo RP, Casola A. Antioxidant Treatment Ameliorates Respiratory Syncytial Virus-induced Disease and Lung Inflammation. Am.J.Respir.Crit Care Med. 2006;174:1361–1369. doi: 10.1164/rccm.200603-319OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Monick MM, Carter AB, Hunninghake GW. Activation of ERK2 by respiratory syncytial virus in A549 cells is linked to the production of interleukin 8. Exp.Lung Res. 2000;26:13–26. doi: 10.1080/019021400269934. [DOI] [PubMed] [Google Scholar]
- Crowe JE, Jr., Williams JV. Immunology of viral respiratory tract infection in infancy. Paediatr.Respir.Rev. 2003;4:112–119. doi: 10.1016/s1526-0542(03)00033-2. [DOI] [PubMed] [Google Scholar]
- Darniot M, Petrella T, Aho S, Pothier P, Manoha C. Immune response and alteration of pulmonary function after primary human metapneumovirus (hMPV) infection of BALB/c mice. Vaccine. 2005;23:4473–4480. doi: 10.1016/j.vaccine.2005.04.027. [DOI] [PubMed] [Google Scholar]
- Domachowske JB, Bonville CA, Gao J-L, Murphy PM, Easton AJ, Rosenberg HF. The chemokine macrophage-inflammatory protein-1alpha and its receptor CCR1 control pulmonary inflammation and antiviral host defense in paramyxovirus infection. Journal of Immunology (Baltimore MD) 2000;165:2677–2682. doi: 10.4049/jimmunol.165.5.2677. [DOI] [PubMed] [Google Scholar]
- Dufour JH, Dziejman M, Liu MT, Leung JH, Lane TE, Luster AD. IFN-gamma-inducible protein 10 (IP-10; CXCL10)-deficient mice reveal a role for IP-10 in effector T cell generation and trafficking. Journal of Immunology. 2002;168:3195–3204. doi: 10.4049/jimmunol.168.7.3195. [DOI] [PubMed] [Google Scholar]
- Einarsson O, Geba GP, Zhu Z, Landry M, Elias JA. Interleukin-11: stimulation in vivo and in vitro by respiratory viruses and induction of airways hyperresponsiveness. Journal of Clinical Investigation. 1996;97:915–924. doi: 10.1172/JCI118514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskildsen S, Hartmann R, Kjeldgaard NO, Justesen J. Gene structure of the murine 2′-5′-oligoadenylate synthetase family. Cell Mol.Life Sci. 2002;59:1212–1222. doi: 10.1007/s00018-002-8499-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esper F, Boucher D, Weibel C, Martinello RA, Kahn JS. Human metapneumovirus infection in the United States: clinical manifestations associated with a newly emerging respiratory infection in children. Pediatrics (Evanston IL) 2003;111:1407–1410. doi: 10.1542/peds.111.6.1407. [DOI] [PubMed] [Google Scholar]
- Espert L, Degols G, Gongora C, Blondel D, Williams BR, Silverman RH, Mechti N. ISG20, a new interferon-induced RNase specific for single-stranded RNA, defines an alternative antiviral pathway against RNA genomic viruses. Journal of Biological Chemistry. 2003;278:16151–16158. doi: 10.1074/jbc.M209628200. [DOI] [PubMed] [Google Scholar]
- Falsey AR, Erdman D, Anderson LJ, Walsh EE. Human metapneumovirus infections in young and elderly adults. Journal of Infectious Diseases. 2003;187:785–790. doi: 10.1086/367901. [DOI] [PubMed] [Google Scholar]
- Fenton M, Sinclair AJ. Divergent requirements for the MAPK(ERK) signal transduction pathway during initial virus infection of quiescent primary B cells and disruption of Epstein-Barr virus latency by phorbol esters. J.Virol. 1999;73:8913–8916. doi: 10.1128/jvi.73.10.8913-8916.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garofalo RP, Haeberle H. Epithelial regulation of innate immunity to respiratory syncytial virus. American Journal of Respiratory Cell and Molecular Biology. 2000;23:581–585. doi: 10.1165/ajrcmb.23.5.f204. [DOI] [PubMed] [Google Scholar]
- Garofalo RP, Sabry M, Jamaluddin M, Yu RK, Casola A, Ogra PL, Brasier AR. Transcriptional activation of the interleukin-8 gene by respiratory syncytial virus infection in alveolar epithelial cells: Nuclear translocation of the RelA transcription factor as a mechanism producing airway mucosal inflammation. Journal of Virology. 1996;70:8773–8781. doi: 10.1128/jvi.70.12.8773-8781.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Plata A, Baron S, Poast JS, Adegboyega PA, Casola A, Garofalo RP. Activity and regulation of alpha interferon in respiratory syncytial virus and human metapneumovirus experimental infections. Journal of Virology. 2005;79:10190–10199. doi: 10.1128/JVI.79.16.10190-10199.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Plata A, Casola A, Garofalo RP. Human metapneumovirus induces a profile of lung cytokines distinct from that of respiratory syncytial virus. Journal of Virology. 2005;79:14992–14997. doi: 10.1128/JVI.79.23.14992-14997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Plata A, Casola A, Suarez G, Yu X, Spetch L, Peeples ME, Garofalo RP. Differential response of dendritic cells to human metapneumovirus and respiratory syncytial virus. Am.J Respir.Cell Mol.Biol. 2006;34:320–329. doi: 10.1165/rcmb.2005-0287OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeberle HA, Casola A, Gatalica Z, Petronella S, Dieterich HJ, Ernst PB, Brasier AR, Garofalo RP. IkappaB Kinase Is a Critical Regulator of Chemokine Expression and Lung Inflammation in Respiratory Syncytial Virus Infection. J.Virol. 2004;78:2232–2241. doi: 10.1128/JVI.78.5.2232-2241.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeberle HA, Kuziel WA, Dieterich HJ, Casola A, Gatalica Z, Garofalo RP. Inducible expression of inflammatory chemokines in respiratory syncytial virus-infected mice: role of MIP-1alpha in lung pathology. Journal of Virology. 2001;75:878–890. doi: 10.1128/JVI.75.2.878-890.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamelin ME, Prince GA, Gomez AM, Kinkead R, Boivin G. Human metapneumovirus infection induces long-term pulmonary inflammation associated with airway obstruction and hyperresponsiveness in mice. J.Infect.Dis. 2006;193:1634–1642. doi: 10.1086/504262. [DOI] [PubMed] [Google Scholar]
- Hamelin ME, Yim K, Kuhn KH, Cragin RP, Boukhvalova M, Blanco JC, Prince GA, Boivin G. Pathogenesis of human metapneumovirus lung infection in BALB/c mice and cotton rats. Journal of Virology. 2005;79:8894–8903. doi: 10.1128/JVI.79.14.8894-8903.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton NH, Mahalingam S, Banyer JL, Ramshaw IA, Thomson SA. A recombinant vaccinia virus encoding the interferon-inducible T-cell alpha chemoattractant is attenuated in vivo. Scand.J.Immunol. 2004;59:246–254. doi: 10.1111/j.0300-9475.2004.01391.x. [DOI] [PubMed] [Google Scholar]
- Hiscott J, Lin R, Nakhaei P, Paz S. MasterCARD: a priceless link to innate immunity. Trends Mol.Med. 2006;12:53–56. doi: 10.1016/j.molmed.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Hiscott J, Pitha P, Genin P, Nguyen H, Heylbroeck C, Mamane Y, Algarte M, Lin R. Triggering the interferon response: The role of IRF-3 transcription factor. J Interferon Cytokine Res. 1999;19:1–13. doi: 10.1089/107999099314360. [DOI] [PubMed] [Google Scholar]
- Indukuri H, Castro SM, Liao SM, Feeney LA, Dorsch M, Coyle AJ, Garofalo RP, Brasier AR, Casola A. Ikkepsilon regulates viral-induced interferon regulatory factor-3 activation via a redox-sensitive pathway. Virology (New York NY) 2006 doi: 10.1016/j.virol.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Jamaluddin M, Garofalo RP, Ogra PL, Brasier AR. Inducible translational regulation of the NF-IL6 transcription factor by respiratory syncytial virus infection in pulmonary epithelial cells. Journal of Virology. 1996;70:1554–1563. doi: 10.1128/jvi.70.3.1554-1563.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jartti T, van den Hoogen BG, Garofalo RP, Osterhaus AD, Ruuskanen O. Metapneumovirus and acute wheezing in children. Lancet. 2002;360:1393–1394. doi: 10.1016/S0140-6736(02)11391-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferies C, Bowie A, Brady G, Cooke EL, Li X, O’Neill LA. Transactivation by the p65 subunit of NF-kappaB in response to interleukin-1 (IL-1) involves MyD88, IL-1 receptor-associated kinase 1, TRAF-6, and Rac1. Molecular & Cellular Biology. 2001;21:4544–4552. doi: 10.1128/MCB.21.14.4544-4552.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CL, Gale M., Jr. CARD games between virus and host get a new player. Trends Immunol. 2006;27:1–4. doi: 10.1016/j.it.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Kahn JS. Human metapneumovirus: a newly emerging respiratory pathogen. Curr.Opin.Infect.Dis. 2003;16:255–258. doi: 10.1097/00001432-200306000-00012. [DOI] [PubMed] [Google Scholar]
- Kahn JS. Epidemiology of human metapneumovirus. Clin.Microbiol.Rev. 2006;19:546–557. doi: 10.1128/CMR.00014-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaisho T, Akira S. Toll-like receptor function and signaling. Journal of Allergy & Clinical Immunology. 2006;117:979–987. doi: 10.1016/j.jaci.2006.02.023. [DOI] [PubMed] [Google Scholar]
- Karin M. The regulation of AP-1 activity by mitogen-activated protein kinase. Journal of Biological Chemistry. 1995;270:16483–16486. doi: 10.1074/jbc.270.28.16483. [DOI] [PubMed] [Google Scholar]
- Karin M. Mitogen-activated protein kinase cascades as regulators of stress responses. Annals of the New York Academy of Sciences. 1998a;851:139–146. doi: 10.1111/j.1749-6632.1998.tb08987.x. [Review] [83 refs] [DOI] [PubMed] [Google Scholar]
- Karin M. The NF-kappa B activation pathway: its regulation and role in inflammation and cell survival. Cancer Journal From Scientific American. 1998b;4(Suppl 1):S92–S99. [Review] [74 refs] [PubMed] [Google Scholar]
- Karin M, Delhase M. JNK or IKK, AP-1 or NF-kappaB, which are the targets for MEK kinase 1 action? Proceedings of the National Academy of Sciences of the United States of America. 1998;95:9067–9069. doi: 10.1073/pnas.95.16.9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Sato S, Yoneyama M, Yamamoto M, Uematsu S, Matsui K, Tsujimura T, Takeda K, Fujita T, Takeuchi O, Akira S. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23:19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, e Sousa Reis, Matsuura Y, Fujita T, Akira S. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature (London) 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. Innate immune recognition of viral infection. Nat.Immunol. 2006a;7:131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. TLR signaling. Cell Death.Differ. 2006b doi: 10.1038/sj.cdd.4401850. [DOI] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat.Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- Kim SH, Han SY, Azam T, Yoon DY, Dinarello CA. Interleukin-32: a cytokine and inducer of TNFalpha. Immunity. 2005;22:131–142. doi: 10.1016/j.immuni.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Laham FR, Israele V, Casellas JM, Garcia AM, Prugent CML, Hoffman SJ, Hauer D, Thumar B, Name MI, Pascual A, Taratutto N, Ishida MT, Balduzzi M, Maccarone M, Jackli S, Passarino R, Gaivironsky RA, Karron RA, Polack NR, Polack FP. Differential production of inflammatory cytokines in primary infection with human metapneumovirus and with other common respiratory viruses of infancy. Journal of Infectious Diseases. 2004;189:2047–2056. doi: 10.1086/383350. [DOI] [PubMed] [Google Scholar]
- Liu P, Jamaluddin M, Li K, Garofalo RP, Casola A, Brasier AR. Retinoic Acid-inducible gene I mediates early antiviral response and toll-like receptor 3 expression in respiratory syncytial virus-infected airway epithelial cells. J.Virol. 2007;81:1401–1411. doi: 10.1128/JVI.01740-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Castro S, Brasier AR, Jamaluddin M, Garofalo RP, Casola A. ROS mediate viral-induced stat activation: role of tyrosine phosphatases. Journal of Biological Chemistry. 2003 doi: 10.1074/jbc.M307251200. [DOI] [PubMed] [Google Scholar]
- Lo MS, Brazas RM, Holtzman MJ. Respiratory syncytial virus nonstructural proteins NS1 and NS2 mediate inhibition of Stat2 expression and alpha/beta interferon responsiveness. J.Virol. 2005;79:9315–9319. doi: 10.1128/JVI.79.14.9315-9319.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas M, Mashimo T, Frenkiel MP, Simon-Chazottes D, Montagutelli X, Ceccaldi PE, Guenet JL, Despres P. Infection of mouse neurones by West Nile virus is modulated by the interferon-inducible 2′-5′ oligoadenylate synthetase 1b protein. Immunol.Cell Biol. 2003;81:230–236. doi: 10.1046/j.1440-1711.2003.01166.x. [DOI] [PubMed] [Google Scholar]
- Ludwig S, Ehrhardt C, Neumeier ER, Kracht M, Rapp UR, Pleschka S. Influenza virus-induced AP-1-dependent gene expression requires activation of the JNK signaling pathway. Journal of Biological Chemistry. 2001;276:10990–10998. [PubMed] [Google Scholar]
- Malathi K, Paranjape JM, Bulanova E, Shim M, Guenther-Johnson JM, Faber PW, Eling TE, Williams BR, Silverman RH. A transcriptional signaling pathway in the IFN system mediated by 2′-5′-oligoadenylate activation of RNase L. Proc.Natl.Acad.Sci.U.S.A. 2005;102:14533–14538. doi: 10.1073/pnas.0507551102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata K, Mibayashi M. The Mx protein that confers the resistance to influenza virus. Nippon Rinsho. 1997;55:2654–2659. [PubMed] [Google Scholar]
- Nam NH. Naturally occurring NF-kappaB inhibitors. Mini.Rev.Med.Chem. 2006;6:945–951. doi: 10.2174/138955706777934937. [DOI] [PubMed] [Google Scholar]
- Nisole S, Stoye JP, Saib A. TRIM family proteins: retroviral restriction and antiviral defence. Nat.Rev.Microbiol. 2005;3:799–808. doi: 10.1038/nrmicro1248. [DOI] [PubMed] [Google Scholar]
- O’Sullivan BJ, MacDonald KP, Pettit AR, Thomas R. RelB nuclear translocation regulates B cell MHC molecule, CD40 expression, and antigen-presenting cell function. Proc.Natl.Acad.Sci.U.S.A. 2000;97:11421–11426. doi: 10.1073/pnas.97.21.11421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onoguchi K, Yoneyama M, Takemura A, Akira S, Taniguchi T, Namiki H, Fujita T. Virus infections activate type I and type III interferon genes through a common mechanism. Journal of Biological Chemistry. 2007 doi: 10.1074/jbc.M608618200. [DOI] [PubMed] [Google Scholar]
- Pachner AR, Bertolotto A, Deisenhammer F. Measurement of MxA mRNA or protein as a biomarker of IFNbeta bioactivity: detection of antibody-mediated decreased bioactivity (ADB) Neurology. 2003;61:S24–S26. doi: 10.1212/01.wnl.0000092361.04511.d0. [DOI] [PubMed] [Google Scholar]
- Pandey S, Agrawal DK. Immunobiology of Toll-like receptors: emerging trends. Immunol.Cell Biol. 2006;84:333–341. doi: 10.1111/j.1440-1711.2006.01444.x. [DOI] [PubMed] [Google Scholar]
- Pazdrak K, Olszewska-Pazdrak B, Liu B, Takizawa R, Brasier AR, Garofalo RP, Casola A. MAP-kinase activation is involved in post-transcriptional regulation of RSV-induced RANTES gene expression. Am J Physiol. 2002 doi: 10.1152/ajplung.00331.2001. 000. [DOI] [PubMed] [Google Scholar]
- Principi N, Bosis S, Esposito S. Human metapneumovirus in paediatric patients. Clin.Microbiol.Infect. 2006;12:301–308. doi: 10.1111/j.1469-0691.2005.01325.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy M, Shi L, Varga SM, Barik S, Behlke MA, Look DC. Respiratory syncytial virus nonstructural protein 2 specifically inhibits type I interferon signal transduction. Virology (New York NY) 2006;344:328–339. doi: 10.1016/j.virol.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Ronni T, Matikainen S, Lehtonen A, Palvimo J, Dellis J, Van Eylen F, Goetschy JF, Horisberger M, Content J, Julkunen I. The proximal interferon-stimulated response elements are essential for interferon responsiveness: a promoter analysis of the antiviral MxA gene. J.Interferon Cytokine Res. 1998;18:773–781. doi: 10.1089/jir.1998.18.773. [DOI] [PubMed] [Google Scholar]
- Ronni T, Matikainen S, Sareneva T, Melen K, Pirhonen J, Keskinen P, Julkunen I. Regulation of IFN-alpha/beta, MxA, 2′,5′-oligoadenylate synthetase, and HLA gene expression in influenza A-infected human lung epithelial cells. Journal of Immunology. 1997;158:2363–2374. [PubMed] [Google Scholar]
- Sato M, Suemori H, Hata N, Asagiri M, Ogasawara K, Nakao K, Nakaya T, Katsuki M, Noguchi S, Tanaka N, Taniguchi T. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity. 2000;13:539–548. doi: 10.1016/s1074-7613(00)00053-4. [DOI] [PubMed] [Google Scholar]
- Sato N, Takahashi N, Suda K, Nakamura M, Yamaki M, Ninomiya T, Kobayashi Y, Takada H, Shibata K, Yamamoto M, Takeda K, Akira S, Noguchi T, Udagawa N. MyD88 but not TRIF is essential for osteoclastogenesis induced by lipopolysaccharide, diacyl lipopeptide, and IL-1alpha. J.Exp.Med. 2004;200:601–611. doi: 10.1084/jem.20040689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz KB. Oxidative stress during viral infection: A review. Free Rad Biol & Med. 1996;21:641–649. doi: 10.1016/0891-5849(96)00131-1. [DOI] [PubMed] [Google Scholar]
- Seger R, Krebs EG. The MAPK signaling cascade. FASEB Journal. 1995;9:726–735. [Review] [99 refs] [PubMed] [Google Scholar]
- Shahabuddin S, Ji R, Wang P, Brailoiu E, Dun N, Yang Y, Aksoy MO, Kelsen SG. CXCR3 chemokine receptor-induced chemotaxis in human airway epithelial cells: role of p38 MAPK and PI3K signaling pathways. Am.J.Physiol Cell Physiol. 2006;291:C34–C39. doi: 10.1152/ajpcell.00441.2005. [DOI] [PubMed] [Google Scholar]
- Smith BT. Cell line A549: a model system for the study of alveolar type II cell function. American Review of Respiratory Disease. 1977;115:285–293. doi: 10.1164/arrd.1977.115.2.285. [DOI] [PubMed] [Google Scholar]
- Smith PL, Lombardi G, Foster GR. Type I interferons and the innate immune response--more than just antiviral cytokines. Mol.Immunol. 2005;42:869–877. doi: 10.1016/j.molimm.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Spann KM, Tran KC, Chi B, Rabin RL, Collins PL. Suppression of the induction of alpha, beta, and lambda interferons by the NS1 and NS2 proteins of human respiratory syncytial virus in human epithelial cells and macrophages [corrected] Journal of Virology. 2004;78:4363–4369. doi: 10.1128/JVI.78.8.4363-4369.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spann KM, Tran KC, Collins PL. Effects of nonstructural proteins NS1 and NS2 of human respiratory syncytial virus on interferon regulatory factor 3, NF-kappaB, and proinflammatory cytokines. J.Virol. 2005;79:5353–5362. doi: 10.1128/JVI.79.9.5353-5362.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson DB, Medzhitov R. Type I interferons in host defense. Immunity. 2006;25:373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Stievano L, Piovan E, Amadori A. C and CX3C chemokines: cell sources and physiopathological implications. Crit Rev.Immunol. 2004;24:205–228. doi: 10.1615/critrevimmunol.v24.i3.40. [DOI] [PubMed] [Google Scholar]
- Takeda K, Akira S. Toll-like receptors in innate immunity. International Immunology (Oxford) 2005;17:1–14. doi: 10.1093/intimm/dxh186. [DOI] [PubMed] [Google Scholar]
- Tian B, Zhang Y, Luxon B, Garofalo RP, Casola A, Sinha M, Brasier AR. Identification of NF-kB dependent gene networks in respiratory syncytial virus-infected cells. Journal of Virology. 2002;76:6800–6814. doi: 10.1128/JVI.76.13.6800-6814.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrence PF. 2-5A-antisense chimeras: inhibitors of respiratory syncytial virus infection. Curr.Opin.Mol.Ther. 1999;1:307–315. [PubMed] [Google Scholar]
- van den Hoogen BG, de Jong JC, Groen J, Kuiken T, de Groot R, Fouchier RA, Osterhaus AD. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat.Med. 2001;7:719–724. doi: 10.1038/89098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JV, Harris PA, Tollefson SJ, Halburnt-Rush LL, Pingsterhaus JM, Edwards KM, Wright PF, Crowe JE., Jr. Human metapneumovirus and lower respiratory tract disease in otherwise healthy infants and children. N.Engl.J Med. 2004;350:443–450. doi: 10.1056/NEJMoa025472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JV, Martino R, Rabella N, Otegui M, Parody R, Heck JM, Crowe JE., Jr. A prospective study comparing human metapneumovirus with other respiratory viruses in adults with hematologic malignancies and respiratory tract infections. J.Infect.Dis. 2005;192:1061–1065. doi: 10.1086/432732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JV, Tollefson SJ, Johnson JE, Crowe JE., Jr. The cotton rat (Sigmodon hispidus) is a permissive small animal model of human metapneumovirus infection, pathogenesis, and protective immunity. J.Virol. 2005;79:10944–10951. doi: 10.1128/JVI.79.17.10944-10951.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JV, Wang CK, Yang CF, Tollefson SJ, House FS, Heck JM, Chu M, Brown JB, Lintao LD, Quinto JD, Chu D, Spaete RR, Edwards KM, Wright PF, Crowe JE., Jr. The role of human metapneumovirus in upper respiratory tract infections in children: a 20-year experience. J.Infect.Dis. 2006;193:387–395. doi: 10.1086/499274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyde PR, Chetty SN, Jewell AM, Schoonover SL, Piedra PA. Development of a cotton rat-human metapneumovirus (hMPV) model for identifying and evaluating potential hMPV antivirals and vaccines. Antiviral Res. 2005;66:57–66. doi: 10.1016/j.antiviral.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Sato S, Hemmi H, Hoshino K, Kaisho T, Sanjo H, Takeuchi O, Sugiyama M, Okabe M, Takeda K, Akira S. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science (Washington DC) 2003a;301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Sato S, Hemmi H, Uematsu S, Hoshino K, Kaisho T, Takeuchi O, Takeda K, Akira S. TRAM is specifically involved in the Toll-like receptor 4-mediated MyD88-independent signaling pathway. Nat.Immunol. 2003b;4:1144–1150. doi: 10.1038/ni986. [DOI] [PubMed] [Google Scholar]
- Yang K, Puel A, Zhang S, Eidenschenk C, Ku CL, Casrouge A, Picard C, von Bernuth H, Senechal B, Plancoulaine S, Al Hajjar S, Al Ghonaium A, Marodi L, Davidson D, Speert D, Roifman C, Garty BZ, Ozinsky A, Barrat FJ, Coffman RL, Miller RL, Li X, Lebon P, Rodriguez-Gallego C, Chapel H, Geissmann F, Jouanguy E, Casanova JL. Human TLR-7-, -8-, and -9-mediated induction of IFN-alpha/beta and -lambda Is IRAK-4 dependent and redundant for protective immunity to viruses. Immunity. 2005;23:465–478. doi: 10.1016/j.immuni.2005.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat.Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- Zachos G, Clements B, Conner J. Herpes simplex virus type 1 infection stimulates p38/c-June N-termianl mitogen-activated protein kinase pathways and activates transcription factor AP-1. J Biol Chem. 1999;274:5097–5103. doi: 10.1074/jbc.274.8.5097. [DOI] [PubMed] [Google Scholar]
- Zapata HJ, Nakatsugawa M, Moffat JF. Varicella Zoster Virus Infection of Human Fibroblast Cells Activates the c-Jun N-terminal Kinase Pathway. J.Virol. 2006 doi: 10.1128/JVI.01470-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Luxon B, Casola A, Garofalo RP, Jamaluddin M, Brasier AR. Expression of RSV-induced chemokine gene networks in lower airway epithelial cells revealed by cDNA microarrays. Journal of Virology. 2001;75:9044–9058. doi: 10.1128/JVI.75.19.9044-9058.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng T, Zhu Z, Wang J, Homer RJ, Elias JA. IL-11: insights in asthma from overexpression transgenic modeling. Journal of Allergy & Clinical Immunology. 2001;108:489–496. doi: 10.1067/mai.2001.118510. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.