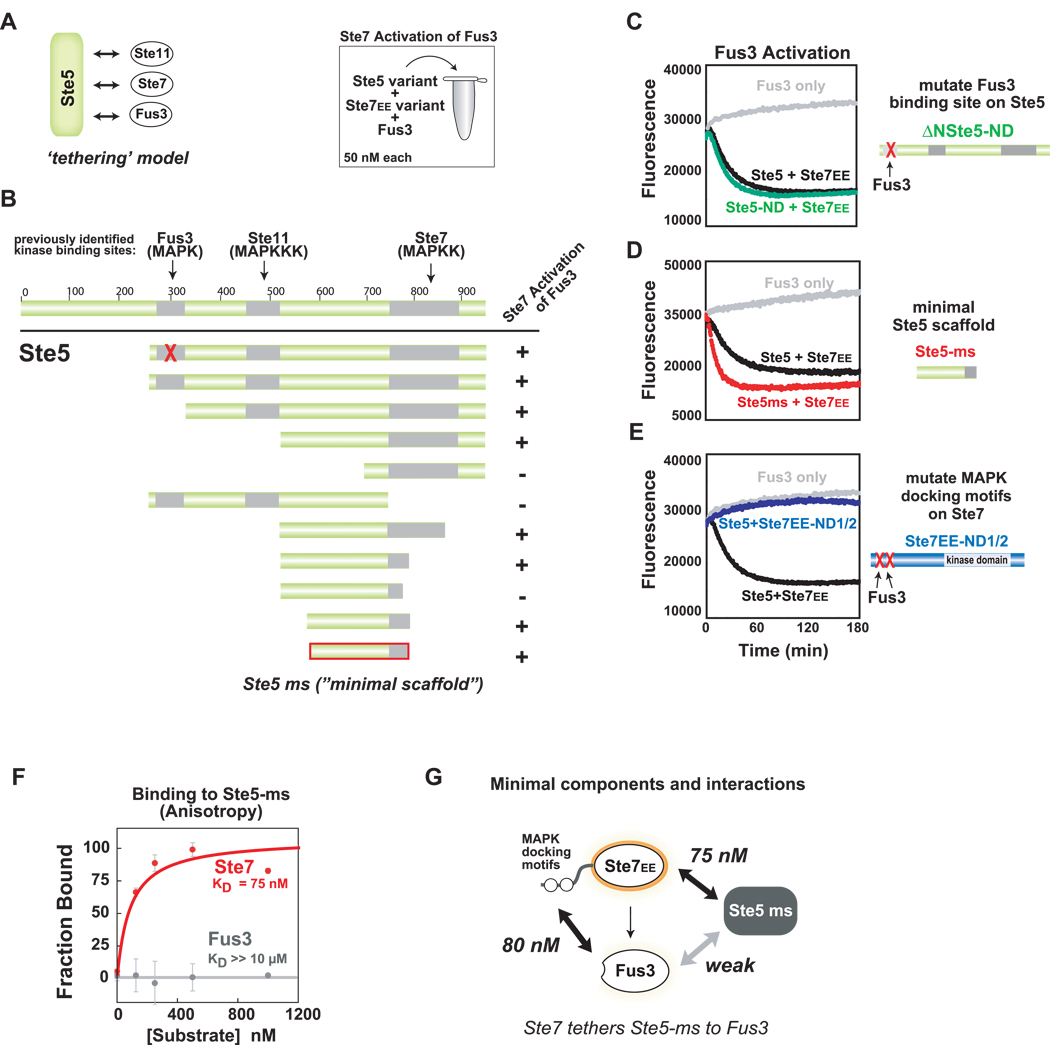
Figure 3. Ste5 contains a novel domain required for Ste7➔Fus3 phosphorylation.
(A) Ste5 is a large protein (917aa) that contains previously identified binding sites for the mating pathway kinases. Canonical tethering model proposes that Ste5 co-localizes three kinases in the mating pathway (Ste11, Ste7, Fus3) to promote signaling. (B) Deletion mapping identifies minimal region of Ste5 required for Ste7EE➔Fus3 phosphorylation in vitro. As in Figure 2, Trulight assay was used to measure Fus3 activation by Ste7EE. Amino acids 593–786 of Ste5 define the ‘minimal scaffold’ domain (Ste5-ms) sufficient to promote Ste7➔Fus3 phosphorylation. (C) Confirmation that the Fus3-binding region (KD = 1 µM) in Ste5 is not required for phosphorylation of Fus3 by Ste7EE. ΔN-Ste5-ND (green curve) is a variant of ΔN-Ste5 (black curve) bearing a mutation in the Fus3 binding region that disrupts interaction with Fus3. For panels C-E all reaction components are at 50 nM. (D) Ste5-ms domain is as active at the larger scaffold protein (ΔN-Ste5). (E) MAPK docking motifs on Ste7EE (KD ~ 100nM) are necessary for Fus3 activation. Mutation of these sites disrupt Ste7➔ Fus3 phosphorylation, even in the presence of Ste5 (purple curve). (F) Ste5-ms binds to Ste7 but not to Fus3. Interactions were measured with fluorescence polarization (anisotropy) using 5nM of fluoroscein-labeled Ste5-ms. (G) Minimal interactions necessary for formation of the Ste5-Ste7-Fus3 signaling complex.