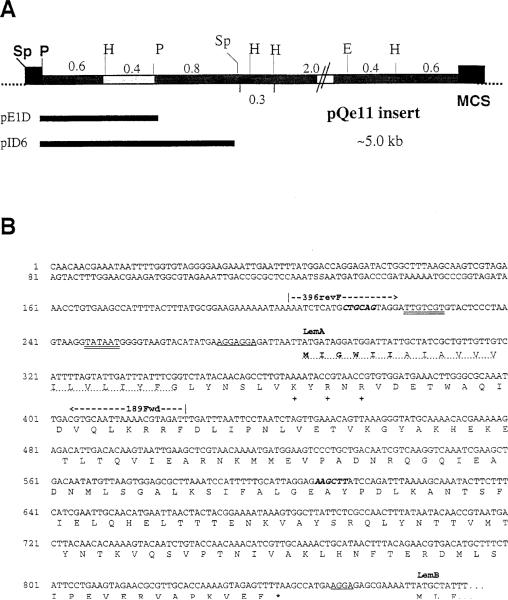
Figure 4. Restriction Map of pQe11 and Partial Nucleotide Sequence of the Putative Lem Operon.
(A) Insert DNA from the pQe11 plasmid was mapped with the indicated enzymes: Sp, SphI; P, PstI; H, HindIII; E, EcoRI. Individual restriction fragments were subcloned into pUC19 and subclones were tested for production of Fr40-like activity. Activity segregated with the highlighted 0.4 kb H–P fragment. Numbers indicate sizes (in kilobases) of the indicated restriction fragments. The pAT29 multiple cloning site (MCS) is represented by closed boxes at each end of the insert. Indicated below the map are the positions of the ~1.2 kb P–P and ~1.8 kb Sp–Sp inserts of subclones pE1D and pID6.
(B) Partial nucleotide sequence from the Listeria insert DNA of pE1D and pID6. Double-underlined bases represent nucleotides that fit consensus sequences for binding of RNA polymerase (–35 and –10). Single underlines designate ribosome-binding sites (rbs). Bold italics indicate the PstI (CTGCAG) and HindIII (AAGCTT) restriction sites, which flank the 0.4 kb fragment to which the Fr40 epitope was mapped (see text). The nucleotide sequence predicts a polycistronic transcript encoding at least two polypeptides (LemA and LemB). Translations for the LemA coding region and the N terminus of LemB are given using the single letter amino acid code. The predicted LemA membrane-spanning domain is indicated by a dotted underline. Positively charged residues used to predict the membrane orientation of LemA are indicated at positions 27, 29, and 31 with a plus symbol. Amino acid residues in bold type represent the sequence of the LemA hexapeptides used in Figure 6, and an asterisk indicates the stop codon for the LemA protein. The coding sequence for LemB continues at least 103 residues and runs into the multiple cloning site (data not shown). The positions of the 5′ (396revF) and 3′ (189Fwd) oligonucleotide primers used in Figure 5 are also shown.