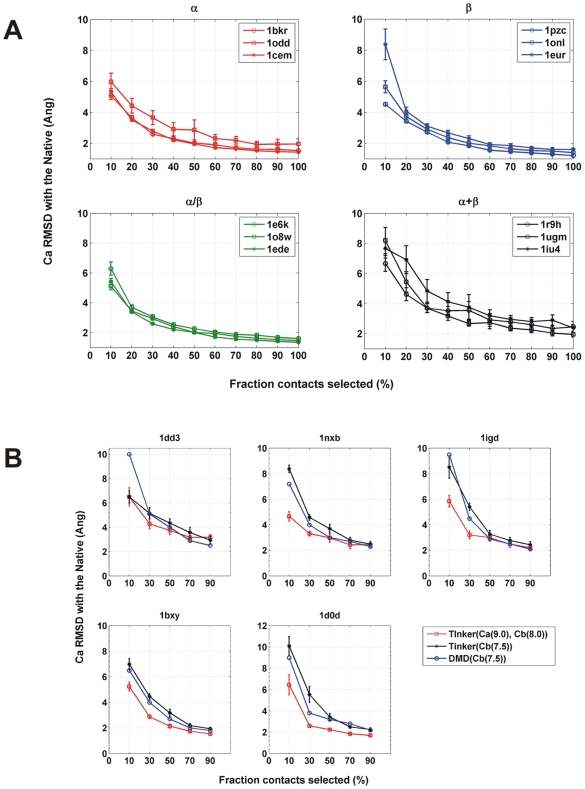
Figure 2. Subsets from random selection.
A Increasing fractions of contacts (from 10% to 100%) are selected at random and reconstructed. Two independent random selections are performed for every fraction and the average Ca RMSD is reported for every protein in a SCOP class. Each class consists of three structures. In each class ‘*’ denotes proteins that are thrice as large as the other two proteins. B The reconstruction accuracies of the random subsets are compared between our method and Chen and co-workers. Five proteins (1dd3, 1nxb, 1igd, 1bxy, 1d0d) are selected from the Chen dataset and the random subsets are generated with (i) our contact definitions Ca 9.0 Å, Cb 8.0 Å (red) (ii) contact definition from Chen et al (Cb 7.5 Å) (black). Subsets from (i) and (ii) are reconstructed with Tinker (iii) The reconstruction accuracy from Chen et al (blue).