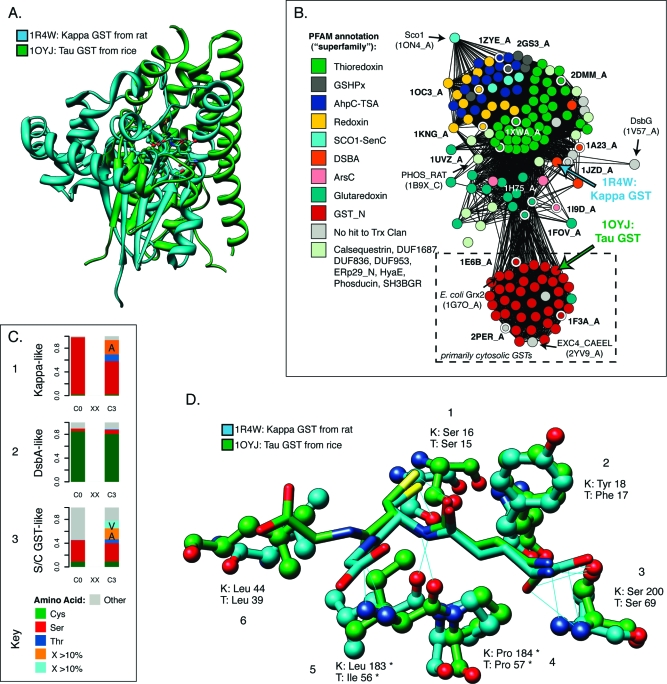
Figure 6.
GST kappa glutathione binding site as an example of convergent evolution. (A) Superposition of the structures of mitochondrial GST kappa (1R4W) and S/C-GST tau (1OYJ) shows that they are very different. While both contain a form of the thioredoxin domain, in kappa, it is interrupted with a large insert before helix 2 (see Figure 4). The alignment superimposes 61 pairs of residues with a 2.17 Å rmsd, all within the Trx domain; six of these amino acids are identical, for a level of sequence identity of ∼10%. The Trx domain in 1OYJ includes ∼80 residues. (B) Within the thioredoxin fold class, GST kappa is structurally distant from the cytosolic GSTs. The structure similarity network shows 159 examples that span the thioredoxin fold. Structures are colored according to their PFAM annotation (see Experimental Procedures), which roughly corresponds to different superfamilies. The locations of 1R4W and 1OYJ are noted. The cytosolic GSTs in Figure 1B are excerpted from this network (dashed box), which is thresholded at a FAST score of 4.5. Edges at this threshold represent alignments with a median rmsd of 2.75 Å across 72 aligned positions. Selected structures (nodes with white borders) in the network are labeled with their PDB entries. (C) The kappa class active site residues are more like cytosolic GSTs than its nearest DsbA-like neighbor class. Bar charts 1−3 show the distributions of amino acids aligning with the catalytic Ser/Cys position in the cytosolic GSTs (C0) and the residue three positions later (C3), commonly termed the CxxC motif. The charts summarize 62, 87, and 538 diverse protein sequences, respectively, from each grouping. (D) Corresponding residues from kappa structure 1R4W and tau cytosolic GST 1OYJ that form binding interactions with glutathione. Predicted H-bonds are shown as thin blue lines. Interactions involving enzyme residues or substructures that are present in all or nearly all variants of the thioredoxin fold are marked with asterisks. Residue pairs are numbered consecutively and discussed in the text.