FIGURE 4.
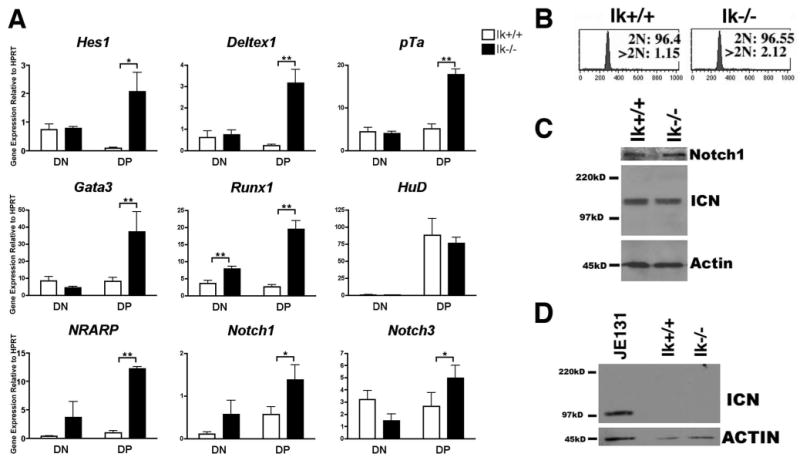
Notch target gene expression is deregulated in primary Ikaros null thymocytes in the absence of deregulated ICN production. A, qRT-PCR analysis was performed using cDNA prepared from sorted thymocyte subsets (double negative, DN; double positive, DP) from Ikaros null (Ik−/−) and wild-type (Ik+/+) mice. Expression levels of the indicated Notch target genes were assessed. Y-axis shows target gene expression normalized to expression of the housekeeping gene, HPRT, in each sample. Values were determined as described in the legend for Fig. 1. Experiments were performed at least three times in duplicate from three independent RNA samples. p values were calculated using paired Student t tests (*, p ≤ 0.05; **, p ≤ 0.02). Error bars represent SEM. Note that there are no significant differences between expression levels in Ik+/+ and Ik−/− DN thymocytes except in the case of Runx1. B, Flow cytometry histograms depicting propidium iodide staining of permeabilized wild-type (Ik+/+) and Ikaros null (Ik−/−) thymocytes. Cells with 2N DNA were designated as being in the G0 or G1 phases of the cell cycle whereas cells with >2N DNA were designated as being in S, G2, or M. C, Western blot analysis was performed using 30 μg of whole cell extract from Ik+/+ and Ik−/− thymuses and Abs that recognize total Notch1 or ICN that has been properly cleaved at val1744. Blots were also probed with anti-actin Abs as a loading control. ICN blot was exposed to autoradiography film for 5 min. D, Western blot analysis of sorted Ik+/+ and Ik−/− DP thymocytes was used to assess the presence of ICN using Abs that recognize properly cleaved ICN. Blots were also probed with anti-actin Abs as a loading control. Whole cell extracts prepared from the JE131 cell line served as a positive control. ICN blot was exposed to autoradiography film for 15 min.
