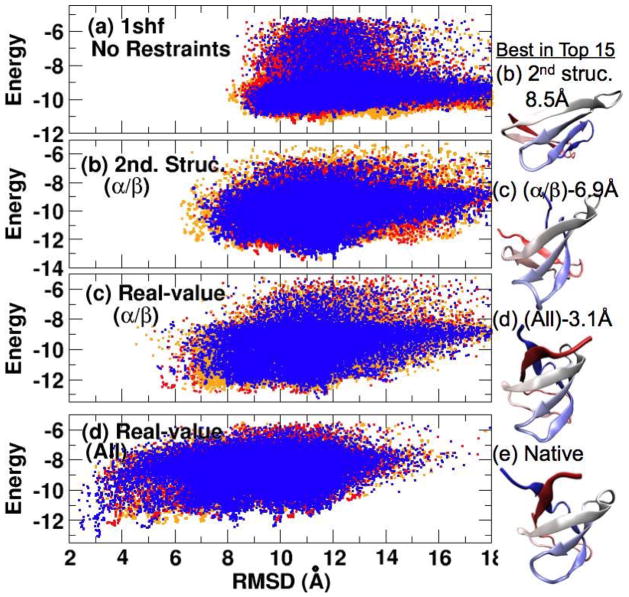
Figure 3.
The effects of various restraints on conformational sampling of 1shf. The energy of all sampled conformations as a function of their RMSD values from the native structure. From top to bottom, conformations are sampled without any restraints (a), with predicted secondary-structure restraints (i.e. restraining around the ideal angles of predicted helical and/or strand residues) (b), with restraints around predicted real-value torsion angles of predicted helical and/or strand residues only (c), and with restraints around predicted torsion angles for all residues (d). The corresponding best structures in top 15 for (b) to (d) are shown in the right panel and the native structure is shown in (e).