Abstract
Vertebrate and invertebrate genomes contain scores of small secreted or transmembrane proteins with two immunoglobulin (Ig) domains. Many of them are expressed in the nervous system, yet their function is not well understood. We analyze here knockout alleles of all eight members of a family of small secreted or transmembrane Ig domain proteins, encoded by the Caenorhabditis elegans zig (“zwei Ig Domänen”) genes. Most of these family members display the unusual feature of being coexpressed in a single neuron, PVT, whose axon is located along the ventral midline of C. elegans. One of these genes, zig-4, has previously been found to be required for maintaining axon position postembryonically in the ventral nerve cord of C. elegans. We show here that loss of zig-3 function results in similar postdevelopmental axon maintenance defects. The maintenance function of both zig-3 and zig-4 serves to counteract mechanical forces that push axons around, as well as various intrinsic attractive forces between axons that cause axon displacement if zig genes like zig-3 or zig-4 are deleted. Even though zig-3 is expressed only in a limited number of neurons, including PVT, transgenic rescue experiments show that zig-3 can function irrespective of which cell or tissue type it is expressed in. Double mutant analysis shows that zig-3 and zig-4 act together to affect axon maintenance, yet they are not functionally interchangeable. Both genes also act together with other, previously described axon maintenance factors, such as the Ig domain proteins DIG-1 and SAX-7, the C. elegans ortholog of the human L1 protein. Our studies shed further light on the use of dedicated factors to maintain nervous system architecture and corroborate the complexity of the mechanisms involved.
NERVOUS systems display an enormous morphological complexity. They are built through the interplay of a plethora of molecular mechanisms that dictate the precise positioning of neuronal cell bodies, axons, dendrites, and synapses. After functional circuits have been established and have begun to operate, nervous systems require dedicated cells and molecules to maintain circuitry structure (Bénard and Hobert 2009). Such active maintenance mechanisms were first described in some molecular and cellular detail in the nematode Caenorhabditis elegans. Its precisely mapped and invariantly structured nervous system allows to probe with unprecedented detail the effect of removing individual cells and genes. Genetic and cellular ablation studies have revealed that a number of diverse Ig domain proteins are required to maintain the precise positioning of axons within fascicles and cell bodies within ganglia (Zallen et al. 1999; Aurelio et al. 2002; Bülow et al. 2004; Sasakura et al. 2005; Wang et al. 2005; Bénard et al. 2006; Pocock et al. 2008). Intriguingly, without these molecules nervous system development is unaffected, yet precise neuroanatomical patterns are lost postdevelopmentally. In other words, within the nervous system, these molecules are dedicated to a role in maintaining the architectural integrity of neuronal circuitry.
Maintenance factors function at specific postembryonic stages (“critical stages”) and they serve to counter disruptive environmental factors, the most notable being mechanical forces exerted onto fascicles and ganglia via movement of the animal. When such movement is inhibited, these displacing forces are absent and consequently maintenance factors are no longer required (Aurelio et al. 2002; Sasakura et al. 2005; Bénard et al. 2006; Pocock et al. 2008).
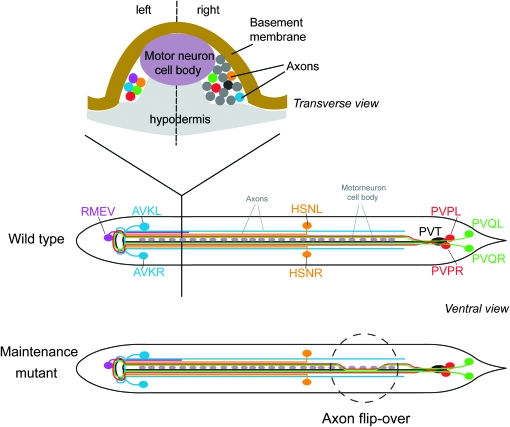
Maintenance factors are required for preserving two prominent structures in C. elegans, axons within the ventral nerve cord and neuronal cell bodies in head ganglia (Bénard and Hobert 2009). The ventral nerve cord of C. elegans is shown schematically in Figure 1. At hatching, the first stage larva (L1 larva) contains only a few axons in the left ventral cord and numerous axons in the right ventral nerve cord. Both cords are separated by a midline composed of motor neuron cell bodies (Figure 1) and at later larval and adult stages by a hypodermal evagination called the hypodermal ridge. The loss of either one of four previously described axon maintenance factors, dig-1, sax-7 (human L1 ortholog), egl-15 (FGF receptor ortholog), and zig-4 results in seemingly identical phenotypes (Aurelio et al. 2002; Bülow et al. 2004; Bénard et al. 2006; Pocock et al. 2008): Axons develop normally in the embryo and appear normally positioned at the early L1 stage, but shortly thereafter become displaced through movement of the animal. sax-7 appears to act autonomously within neurons (Pocock et al. 2008) and dig-1 cell nonautonomously from muscle (Bénard et al. 2006); both factors also are required to maintain cell body position in head ganglia. In contrast, egl-15 and zig-4 are only required to maintain axon position in the ventral nerve cord; to fulfill this role, egl-15 acts in the hypodermis that underlies that ventral nerve cord (Bülow et al. 2004) and zig-4 appears to be secreted from PVT, a unilateral neuron that extends an axon along the ventral cord. PVT ablation phenocopies the loss of zig-4 (Aurelio et al. 2002). How all these factors interact is at present not understood.
Figure 1.—
Overview of the ventral nerve cord in C. elegans. Nervous system architecture of C. elegans has been described in detail by White et al. (1986).
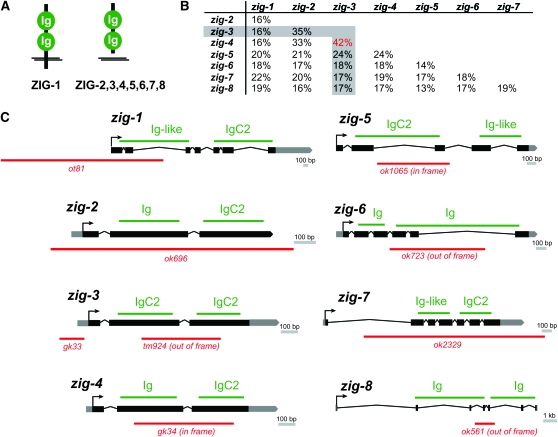
To better understand the maintenance of neuronal architecture, we set out to identify additional maintenance factors. The previously identified maintenance factor ZIG-4 is a member of a family of eight ZIG proteins, which are all defined by their common domain architecture. Each protein is composed of two Ig domains (hence the name “Zwei Ig Domänen”) and is predicted to be either secreted (ZIG-2–ZIG-8) or cell surface associated on the basis of the presence of a transmembrane domain (ZIG-1) (Figure 2A). On the primary sequence level, C. elegans ZIG proteins are diverse (Figure 2B) and apparently fast evolving, as evidenced in the distinct complement of ZIG proteins apparent in the related nematodes C. briggsae and C. remanei (www.wormbase.org). Fly and vertebrate genomes also contain scores of small secreted or transmembrane two-Ig domain proteins, which, however, bear little primary sequence similarity to ZIG proteins (Rougon and Hobert 2003).
Figure 2.—
zig genes, proteins and mutant alleles. (A) Schematic structure of ZIG proteins. (B) Sequence comparison of ZIG proteins. Percent identity was calculated from alignment done with T-coffee (Notredame et al. 2000). (C) zig mutant alleles. The type of Ig domains are indicated as identified by SMART (http://smart.embl-heidelberg.de/).
Aside from zig-4, no other zig gene has previously been functionally characterized. In this article we address the question of whether other zig family members may, like zig-4, also be involved in maintaining nervous system structure. This seemed a particularly attractive possibility as many family members are expressed by the same neuron in the ventral cord, PVT, which we found to be required for maintaining fascicle structure (Aurelio et al. 2002). Moreover, since zig-4 mutants affect the maintenance of only a subset of axons in the ventral nerve cord, the question arose whether other zig genes affect other axons. Lastly, other maintenance factors, such as dig-1 or sax-7 affect cell body position, prompting the question of whether other ZIG proteins affect the maintenance of cell body position as well. To address these questions we undertook a systematic functional analysis of all zig family members using genetic deletion alleles. We report the result of these studies in this article.
MATERIALS AND METHODS
Strains:
Strains were maintained as described (Brenner 1974). A list of all strains used in this study is provided in the supporting information, File S1.
All deletion alleles were generated by the C. elegans knockout consortia through PCR screening of mutagen-induced deletion libraries, with the exception of zig-1(ot81). The zig-1(ot81) deletion allele was generated by spontaneous imprecise excision of a Tc1 transposable element from the strain cxTi6127; mut-7(pk204) dpy-18(e364) provided by Laurent Segalat. This strain carries a Tc1 transposon insertion at position 3301 in the cosmid K10C3. In brief, 10 adult worms were placed on 150 plates and grown at 20° until food was exhausted. Half of the worms were then collected from each plate, lysed, and screened for deletion products by PCR. Positive plates were chunked in four parts and retested by PCR. Finally, single worms were picked from positive plates to isolate a homozygous deletion mutant. Sequencing of the zig-1(ot81) breakpoints revealed a 2281-bp deletion from position 3282–5563 in K10C3, which deletes the first two exons, the entire first intron, and part of the second intron of zig-1. All alleles were outcrossed with N2 at least three times.
Genotyping of zig gene alleles throughout various genetic crosses was carried out by PCR, as their deletion results in no obvious morphological or behavioral phenotypes. PCR was done on genomic DNA preparations, generated by lysing mixed population of worms grown on agar plates using a standard protocol (Fay and Bender 2008). Primers used for PCR are listed in Table S1.
The genes zig-3 and zig-4 map only 0.06 cM apart on LGX (<5 kb apart). To minimize the effort of screening by PCR for a recombinant between these two genes, we used the genetic markers lon-2(e678) and unc-115(mn481) flanking zig-3 and zig-4. On LGX, lon-2 is located at −6.7 cM, zig-4 at −1.00 cM, zig-3 at −0.94 cM, and unc-115 at 1.88 cM. We first generated the two starting strains, lon-2(e678) zig-4(gk34) and zig-3(tm924) unc-115(mn481), by genetic recombination from heterozygous worms of genotypes lon-2unc-18/zig-4 and dpy-6unc-115/zig-3, respectively. We then crossed lon-2zig-4/00 males into zig-3unc-115 hermaphrodites to obtain heterozygous hermaphrodites lon-2zig-4/zig-3unc-115. From these heterozygous F1 worms, we picked a total of 800 Lon or Unc F2 worms. Two hundred eighty-six of these Lon or Unc F2 are expected to carry a recombinant chromosome of the type lon-2unc-115, among which only 2 would be lon-2zig-4zig-3unc-115, brought about by a recombination event taking place between zig-3 and zig-4. We homozygosed lon-2unc-115 chromosomes by singling F3 worms that were Lon and Unc. We lysed worms and analyzed by PCR for the presence of both zig-3 and zig-4 mutant and wild-type alleles to detect recombinants occurring between these two genes. We obtained one recombinant lon-2zig-4zig-3unc-115. We then proceeded to get a zig-4zig-3 strain devoid of the intermediate markers lon-2 and unc-115. We first removed unc-115 by picking Lon-non-Unc recombinant F2 worms from heterozygous F1 lon-2zig-4zig-3unc-115/+. We homozygosed lon-non-unc chromosomes by singling Lon F3 worms. We lysed and analyzed them by PCR to select those retaining zig-4zig-3. We next removed the marker lon-2 by picking non-Lon F2 worms from heterozygous F1 hermaphrodites of the genotype lon-2zig-4zig-3/oxIs12 generated by crossing oxIs12/0 males into lon-2zig-4zig-3. oxIs12 is a dominant transgene integrated at ∼2 cM of LGX that drives gfp in GABAergic neurons. We selected F3 broods with 75% oxIs12 and no Lon worms. We next homozygosed the new zig-4zig-3 chromosome, devoid of lon-2, by selecting for non-oxIs12 worms. We confirmed by PCR the homozygous presence of zig-3(tm924) and zig-4(gk34).
Transgenes:
Transgenes generated for rescue of mutant phenotypes are listed in File S1, with concentration of injected DNA indicated. Reporter transgenes to score neuroanatomy are listed in Table 1. They were crossed with zig deletion alleles and the resulting strains genotyped for the presence of the zig deletion alleles by PCR (Table S1). One panneuronal reporter transgene, otIs173, was specifically created for this study. pCB101=F25B3.3∷DsRed2 was generated by cloning a BamHI–ApaI fragment containing DsRed2 under the F25B3.3 promoter. pCB101 was injected at 50 ng/μl along with ttx-3∷gfp at 75 ng/μl. An extrachromosomal array was integrated by gamma irradiation, by conventional protocols, yielding otIs173. The strain bearing otIs173 was outcrossed 10 times. otIs173 maps to LGIII. Another reporter transgene, zig-3 short prom∷gfp, was generated by PCR fusion (Hobert 2002) (PCR primers see “DNA constructs” in File S1).
TABLE 1.
Axon anatomy transgenic markers
| Transgene name | Reporter construct | Cell(s) labeled | Reference |
|---|---|---|---|
| oyIs14 | sra-6∷gfp | PVQ, ASH, ASI | Sarafi-Reinach et al. (2001) |
| hdIs26, hdIs29 | sra-6∷DsRed2 odr-2∷cfp | PVQ, ASH, ASI PVP, and other head and VNC neurons | Hutter (2003) |
| oxIs12 | unc-47∷gfp | RMEV and all other GABAergic neurons | McIntire et al. (1997) |
| juIs8 | unc-25∷gfp | RMEV and all other GABAergic neurons | Jin et al. (1999) |
| zdIs13 | tph-1∷gfp | HSN, ADF, NSM | Clark and Chiu (2003) |
| bwIs2 | flp-1∷gfp | AVK only | Much et al. (2000) |
|
otIs173 |
F25B3.3∷DsRed2 |
Panneuronal |
This work |
DNA constructs:
DNA constructs were generated by standard cloning procedures or by PCR fusion (Hobert 2002). A list of all constructs and primer sequences can be found in File S1.
Scoring of neuroanatomy:
Axon and cell body position were scored with reporter transgenes listed in Table 1. Unless otherwise indicated, animals were grown at 20° and scored using a Zeiss Axioplan 2 microscope. Axons of the ventral nerve cord were scored in freshly hatched L1 larvae as well as in young adults. To obtain freshly hatched L1 larvae, embryos were picked and allowed to hatch and develop no longer than 30 min posthatching. Young adults that have just molted from L4, have a slightly protruding vulva and no embryos in their uteri. Cell body position was examined in 3- to 5-day-old adults. All phenotypes were scored as percentage of animals defective and results are shown with error bars representing the standard error of proportion. Statistical significance was calculated using the z-test to compare the proportion of abnormal animals of two genotypes. When using the same control for multiple comparisons, the P-value was multiplied by the total number of comparisons.
Laser ablation:
Laser ablations were performed as previously described (Bargmann and Avery 1995) using a Photonics dye laser (LSI VSL-337) attached to a Zeiss Axioplan 2 microscope equipped with Nomarski, and fluorescence optics were used for microscopy. Freshly hatched L1 worms were mounted in a drop of 10 μm sodium azide solution on a 5% agarose pad, sealed between a cover slip, and observed directly using Nomarski and fluorescence microscopy to identify the targeted cells. We used the otEx2419gcy-1∷gfp array (Ortiz et al. 2006) to visualize and ablate PVT in wild-type and zig-4zig-3 double mutant background. For ablation of PVQL, PVQR, PVPL, and PVPR, reporter strains oyIs14 and hdIs26 were used, respectively. Cells were irradiated for ∼3 sec in the nucleus. Increased cytoplasmic movements were often observed and in many cases the nucleus was observed to break down. Animals were recovered promptly and allowed to develop to the young adult stage at 20°, when their neuroanatomy was examined. Successful ablation of the neuron was assessed by the absence of gfp fluorescence at the adult stage.
RESULTS
Analysis of zig gene knockout alleles:
Of the eight zig genes only one, zig-4, was previously analyzed through gene deletion analysis (Aurelio et al. 2002). Through generating deletion alleles by either transposon mobilization (zig-1) or deletion library screening, conducted by the C. elegans knockout consortia (zig-2, -3, -5, -6, -7, and -8), we have obtained knockout alleles for each of the so far uncharacterized, seven remaining zig genes in C. elegans. Each allele is a predicted molecular null allele (Figure 2C) and none affects viability or produces any other readily visible morphological or behavioral phenotype. Given the zig-4 precedent and the expression of at least six of the eight zig genes in the PVT neuron of the ventral nerve cord (Aurelio et al. 2002), we analyzed ventral nerve cord architecture in all zig mutant animals. In addition, we also examined the correct position of neuron soma; correct maintenance of soma position is controlled by other previously identified maintenance factors (Zallen et al. 1999; Sasakura et al. 2005; Bénard et al. 2006). This neuroanatomical analysis was conducted by crossing a series of chromosomally integrated gfp reporters that allow the visualization of individual neuron types in all zig mutant backgrounds (Table 1). No defects were observed for any zig mutants in head neuron position (data not shown), yet we found that zig-3 mutants, but no other single zig mutants, display position defects of individual and specific sets of axons in the ventral nerve cord (Table 2; Figure 3). Two different zig-3 alleles display similar defects and defects can be rescued by reintroducing wild-type copies of the zig-3 genomic locus (Figure 4).
TABLE 2.
Phenotypes of zig mutants
| % animals with axon flip-over defects of the following neuron classes |
|||||
|---|---|---|---|---|---|
| Genotype | PVQ | PVP | RMEV | HSN | AVK |
| Wild type | 7% | 6.0% n = 285a | 1% n = 118c | 4.7% | 1% |
| n > 400 | 7.9% n = 178b | 1% n = 71d | n = 169 | n = 120 | |
| zig-1(ot81) | 8% | 4.9% | 0% | 3.2% | 2.3% |
| n = 177 | n = 122a | n = 86c | n = 126 | n = 87 | |
| zig-2(ok696) | 8% | 4% | 1% | 1.8% | 0% |
| n = 187 | n = 85a | n = 78c | n = 112 | n = 104 | |
| zig-3(tm924) | 26% | 20% | 3.3% | 0% | 1% |
| n = 201 | n = 160a | n = 120d | n = 130 | n = 196 | |
| zig-4(gk34) | 24% | 22% | 1.6% | 3.8% | 1% |
| n = 189 | n = 144a | n = 122d | n = 131 | n = 105 | |
| zig-4(gk34) zig-3(tm924) | 26% | 24.6% | 4.7% | 10% | 0% |
| n = 299 | n = 240a | n = 129d | n = 248 | n = 85 | |
| zig-5(ok1065) | 9% | 4.4% | 0% | 6.3% | 0% |
| n = 194 | n = 90b | n = 96c | n = 175 | n = 93 | |
| zig-6(ok273) | 9% | 5.5% | 2.6% | 7% | 0% |
| n = 166 | n = 127a | n = 115d | n = 116 | n = 106 | |
| zig-7(ok2329) | 5% | 9.2% | 0% | 4.2% | 0% |
| n = 100 | n = 130a | n = 94c | n = 118 | n = 100 | |
| zig-8(ok561) | 9% | 11% | 0% | 6.5% | 0% |
|
n = 193 |
n = 168a |
n = 85c |
n = 138 |
n = 70 |
|
Defects that are statistically different (P < 0.05) from wild type are in boldface type. Transgenes used to visualize axon anatomy: PVQ, oyIs14; PVP, hdIs26a or hdIs29b; RMEV, oxIs12c or juIs8d; HSN, zdIs13; AVK, bwIs2. See Table 1 for more information on these transgenes.
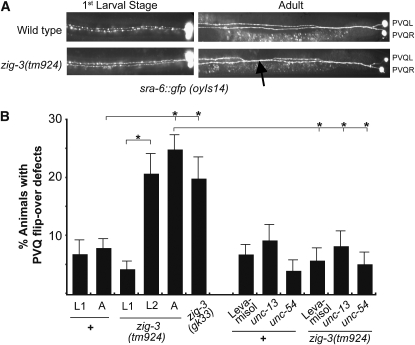
Figure 3.—
zig-3 mutant phenotype. (A) PVQ flip-over defects observed with the oyIs14 reporter transgene. Most (>90%) of all axon flip-over defects observed in zig-3 (or zig-4) mutant animals occur in the posterior half of the animal. (B) Quantification of zig-3 mutant defects at different developmental stages and under different experimental conditions. Proportions of different animal populations were compared using the z-test. *P < 0.05.
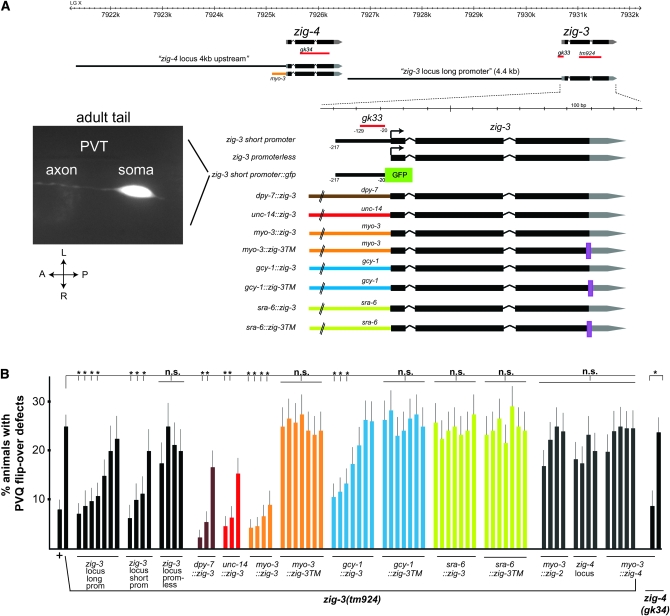
Figure 4.—
Expression and focus of action of zig-3. (A) zig-3 and zig-4 genomic locus and constructs used to test for zig-3 and zig-4 rescuing activity and reporter gene analysis. Two independent transgenic lines showed the same gfp expression pattern. A young adult animal is shown. (B) Quantification of rescue data. Note that we have previously shown that overexpression of zig-4, using again the myo-3 promoter, causes PVQ axon outgrowth defects (Aurelio et al. 2003). In the experiments shown here myo-3∷zig-4 was injected at lower concentration, which did not result in axon outgrowth defects. One of the myo-3∷zig-4 lines generated in the zig-3(tm924) background was transferred into zig-4(gk34) to verify its rescuing ability of the zig-4 mutant defects. Proportions of different animal populations were compared using the z-test. *P < 0.05; NS: P > 0.05.
The axonal defects in zig-3 mutants are postdevelopmental in nature, as they are not observed in freshly hatched larvae (examined at <30-min posthatching, Figure 3). That is to say, axons that have migrated along the ventral nerve cord during embryogenesis appear normal at the early L1 stage, but during later larval stages become displaced across the ventral midline. This displacement results from mechanical stress as it can be suppressed through immobilizing worms either pharmacologically or through incapacitating locomotion genetically (Figure 3). We conclude that the normal function of zig-3 is to prevent locomotion-induced axon displacement to occur.
Genetic interactions of zig-3:
The zig-3 mutant defects are very similar to those seen in zig-4 mutants, in terms of appearance, penetrance, and cellular specificity (Table 2). We generated a double null mutant of zig-3 and zig-4 and find that those double mutant animals display similar phenotypes compared to those observed in the respective single null mutants (Figure 5A). The failure to enhance null mutant phenotypes usually is an indication of two genes acting in the same genetic pathway. In the case of ZIG-3 and ZIG-4, acting in a similar pathway may mean that both proteins may act together in a complex, as many other Ig proteins do (Barclay et al. 1997). We note that failure to enhance each other's null phenotype is not due to a “ceiling effect,” as other mutant combinations of maintenance factors display much stronger phenotypes (C. Bénard and O. Hobert, unpublished data).
Figure 5.—
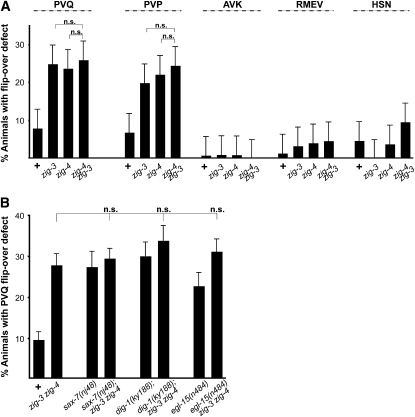
Genetic interaction data. Genetic interaction tests of zig-3(tm924), zig-4(gk34) and other maintenance mutants. “+” = wild-type strain, “zig-3 zig-4” = zig-3(tm924) zig-4(gk34). See Table 1 for reporters used. All animals were scored as young adults. Proportions of different animal populations were compared using the z-test. NS: P > 0.05.
The zig-3 single mutant, the zig-4 single mutant, and a zig-4zig-3 double mutant also do not enhance the VNC axon flip-over phenotype of other axon maintenance factors, namely dig-1, sax-7, and egl-15 (Figure 5B). This indicates that all these genes may act in one common pathway.
A regulatory allele of zig-3 affects a neuronal cis-regulatory sequence:
Where does zig-3 act to affect axon maintenance? We had previously reported that 4.4 kb of 5′ upstream regulatory sequences of the zig-3 locus drive gfp reporter gene expression in PVT and a few other neurons and that ablation of PVT displays axon maintenance defects similar to those that we now observe for zig-3 (Aurelio et al. 2002). One of the two zig-3 alleles, gk33, is a regulatory allele that further bolsters the hypothesis of a neuronal focus of action of zig-3. gk33 animals carry a 109-bp deletion that does not affect the protein-coding part of zig-3 but deletes sequences from 129 to 20 bp upstream of the predicted start codon of zig-3 (Figure 2). A genomic fragment of the zig-3 locus, termed “zig-3 short promoter,” which contains exons, introns, the 3′-UTR, and those 5′ upstream sequences deleted in gk33 (plus a few dozen nucleotides) rescues the zig-3 mutant phenotype (Figure 4). Removal of these 5′ sequences from this rescuing construct abolishes rescuing activity (Figure 4) (zig-3 promoterless). When fused to gfp, the short 5′ promoter shows expression in a small number of cells with the only neuron in the ventral nerve cord being PVT (Figure 4).
zig-3 gene activity can be ectopically provided from several different cell types but normally appears to be provided by PVT:
Consistent with a focus of action of zig-3 in PVT, we find that expression of zig-3 under control of the gcy-1 promoter, which is active in PVT, the gut, and a few head neurons (Ortiz et al. 2006), also rescues the zig-3 mutant phenotype (Figure 4). If we tag the zig-3 locus with a transmembrane domain from the pat-3 gene, and express it under control of the gcy-1 promoter, rescuing ability is abolished, suggesting that zig-3 must be secreted to exert its function (Figure 4).
Secreted zig-3 can function from several distinct but not all cellular sources. If we express zig-3 under control of the myo-3 promoter, which is exclusively active in body wall muscle cells that abut the ventral nerve cord (Okkema et al. 1993), we observe full rescue of the mutant phenotype (Figure 4). zig-4 expressed under control of the myo-3 promoter can also rescue the zig-4 mutant phenotype (Figure 4). The rescuing activity is abolished, however, when we append a transmembrane domain to the myo-3-driven zig-3 gene, pointing to the need for ZIG-3 to be secreted and diffusible (Figure 4).
Expression under the control of a promoter that is active in all neurons (unc-14) (Ogura et al. 1997) and under a promoter that is active exclusively in hypodermal cells that underlie the ventral nerve cord (dpy-7) (Gilleard et al. 1997) also rescues the mutant phenotype (Figure 4). In contrast, zig-3 expression under the control of the sra-6 promoter, which is active only in the PVQ neurons in the ventral nerve cord, does not rescue the mutant phenotype (Figure 4); this is somewhat surprising and may relate to aberrant expression levels provided by this promoter or by the inability of the sra-6 expressing cells to secrete functional ZIG-3. We conclude that as long as zig-3 is secreted it can be supplied from different, but not necessarily all cell types. Similarly, zig-4 can also function when heterologously supplied from distinct cell types (Aurelio et al. 2002).
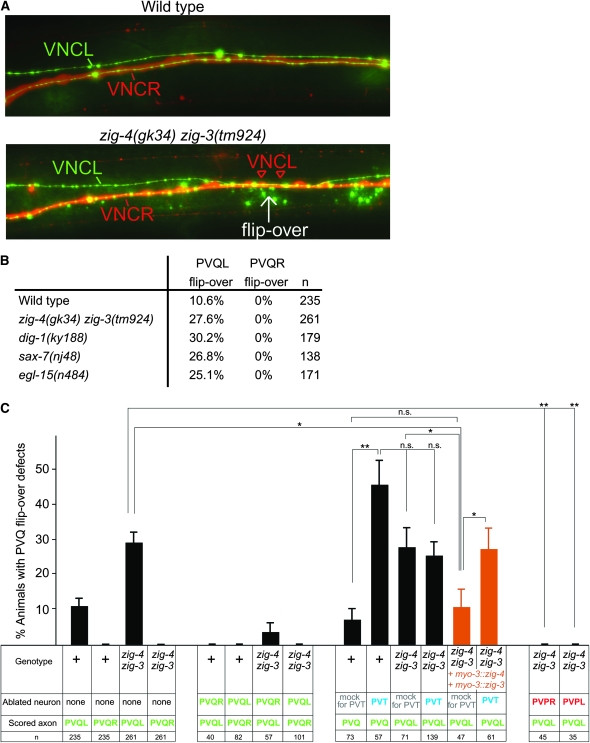
Several additional lines of evidence besides PVT expression and heterologous rescue with the gcy-1 promoter point to a focus of action of zig-3, and also of zig-4, in PVT. Previous laser ablation studies (Aurelio et al. 2002), which we recapitulate here (Figure 6C), show that postembryonic (early L1 stage) ablation of PVT phenocopies the axon maintenance defects observed in zig-3 and zig-4 mutant animals. If zig-3 and zig-4 acted from cells other than PVT, then the defects observed upon PVT ablation should be additive to those seen in zig-3zig-4 double mutants; if they act from PVT, the defects should not be any more severe. We find that the frequency of the flip-over defects observed upon postembryonic PVT ablation are not any more severe if done in a zig-4zig-3 double mutant background, consistent with PVT being the normal source of zig-3 and zig-4 (Figure 6C).
Figure 6.—
Cellular analysis of axon midline flip-over defects. (A) Midline flip-overs are all-or-none defects as assessed by colabeling the entire left and right ventral nerve cord fascicle with a panneuronal rfp marker (otIs173) and the PVQ neurons with a gfp marker (oyIs14). Young adult animals are shown. (B) PVQ axon flip-overs (scored in young adults with the oyIs14 and otIs173 transgenes) always occur from the left into the right fascicle, as scored in different maintenance factor mutant backgrounds. This also holds for the background level of flip-over in a wild-type genetic background. (C) Laser ablation data. “+” = wild-type strain; “zig-4 zig-3” = zig-3(tm924) zig-4(gk34). For the PVT ablation experiments, a gcy-1∷gfp transgene was placed into the genetic background to visualize PVT. PVQ axons and soma were visualized with oyIs14 and PVP axons and soma with hdIs26. Proportions of different animal populations were compared using the z-test. *P < 0.05; **P < 0.01; and NS: P > 0.05.
PVT also has zig-3/zig-4-independent roles:
The data described above suggests that PVT provides zig-3 and zig-4 function. Is this the only way in which PVT affects axon maintenance? If so, then PVT would be superfluous if we provided zig-3 and zig-4 from heterologous cells. We tested this in the following manner: As we described above, zig-3 and zig-4 gene function can be provided using the heterologous myo-3 promoter. We corroborated that this is the case in a zig-4zig-3 double mutant; a transgenic array that contains both myo-3∷zig-3 and myo-3∷zig-4 can indeed rescue the zig-4zig-3 double mutant phenotype (Figure 6C). In those animals, zig-3 and zig-4 are therefore not provided from their normal cellular source (PVT) but from an ectopic source, resulting in phenotypically wild-type animals. When we ablate PVT in those transgenic animals, we observe axon maintenance defects again (Figure 6C). This indicates that PVT does more than just providing zig-3 and zig-4 function. Indeed we have found that a simultaneous deletion of two other PVT-expressed and apparently redundantly acting zig genes also results in axon maintenance defects (C. Bénard and O. Hobert, unpublished data), suggesting that PVT does provide multiple zig genes that act nonredundantly to control axon maintenance at the midline.
zig gene action is highly sequence specific:
Even though all ZIG proteins share a two-Ig domain architecture, they are very diverse at the primary sequence level, with some ZIG proteins displaying as little as 13% primary sequence identity (Figure 2). In relative terms, ZIG-3 and ZIG-4 are the most closely related ZIG proteins, not only by primary sequence (42% identical), but also by conserved intron/exon structure and chromosomal location. The two genes directly neighbor one another (Figure 4A). zig-3 and zig-4 therefore likely arose by local duplication that not only duplicated protein-coding sequences but also cis-regulatory control regions as the 5′ regulatory region of each gene drives expression in PVT independently (Aurelio et al. 2002). In spite of these similarities, a loss of either zig-3 or zig-4 results in a similar mutant phenotype, not further enhanced in the double mutant. This already suggests strongly that the two genes do not act in a redundant manner. To test for gene dosage effects, we asked whether the zig-3 mutant phenotype can be rescued by overexpressing the zig-4. Multicopy transgenic arrays show that this is not the case (Figure 4B). Moreover, using the myo-3 driver, we find that only myo-3∷zig-3, but not myo-3∷zig-4, can rescue the zig-3 mutant phenotype. Similarly, the zig-2 gene, which is the second closest homolog to zig-3 (35% primary sequence identity; Figure 2B), is also not able to rescue the zig-3 mutant phenotype when driven under control of the myo-3 promoter (Figure 4B).
Mechanistic aspects of the axon flip-over defect:
We examined the axon flip-over defect observed in zig-3, as well as in other axon maintenance mutants in more detail, with the specific goal of defining whether axons flip into the opposite fascicle of the cord or whether they stray off into more intermediate midline locations. In previous reports on axon maintenance defects (Aurelio et al. 2002; Bénard et al. 2006; Pocock et al. 2008), we had only scored axon flip-over defects if the left and right axons made contact with one another (as observed on the light microscopy level), leaving the possibility that more subtle mispositioning defects, in which only one axon moved onto the midline (rather than into the opposite fascicle), were not reported. Moreover, we had previously surmised that axon flip-over defects in zig-4 show no directionality in terms of whether an axon crosses from the left to the right fascicle or vice versa (Aurelio et al. 2002), but without more fine-grained anatomical resolution, this conclusion was only tentative. To analyze these issues in more detail, we constructed and used a panneuronal rfp marker transgene (otIs173) to unambiguously distinguish the right from the left fascicle, thereby generating a clear reference point. Animals in which the entire left and right fascicles are labeled in red and the PVQ axons are specifically labeled in green reveal that PVQ axon flip-overs observed in various maintenance factor mutant backgrounds are all-or-nothing defects (Figure 6A). No intermediate positions of flipped axons are observed. Curiously, we noted that it is exclusively the axons of the less populated left fascicle that flip over the midline (i.e., PVQL) but never from the much thicker right fascicle into the left fascicle (Figure 6, A and B).
As a flipped-over axon always appears to make contact with its contralateral analog, at least on the light microscopy level, we asked whether PVQR is required for the manifestation of the PVQL flip-over defect observed in zig mutants. We find this to be indeed the case. When we ablate PVQR postembryonically (early L1 stage) in zig-4zig-3 double mutants, we observe that the flip-over of the axon of PVQL into the right VNC that usually occurs is completely suppressed (Figure 6C). These results corroborate previously conducted PVT/PVQR double ablation experiments (Aurelio et al. 2002). Unexpectedly, postembryonic ablation of either of two embryonically generated PVP neurons, whose axons closely fasciculate with the PVQ axons (see Figure 1 for schematic), also completely suppresses the PVQ axon flip-over defects of zig-4zig-3 mutants (Figure 6C). While the requirement of the PVP axon in the right cord (PVPL) may be similar to the requirement of the right PVQ axon (both may actively recruit and/or stabilize PVQL during/after a midline flip-over event), the suppression of the PVQL flip by ablating its directly neighboring left cord-residing PVPR axon is less obvious to explain. Apparently, if maintenance mechanisms are disabled, a latent activity of PVPR pushes the PVQL axon away into the opposite cord; eliminating PVPR removes this impetus and PVQL remains in the left fascicle.
DISCUSSION
We have identified a new maintenance factor, zig-3, which is required to stabilize axonal architecture at the ventral midline. We have identified the role of this factor in the course of a comprehensive mutant analysis of an entire family of small secreted Ig domain proteins. Most of them do not display a phenotype in the cellular context that we examined, but we surmise that they may have functions in other, as yet nonexamined cell types or that they may act in a redundant manner. A preliminary analysis of double mutant combinations indeed points to such a redundant scenario (C. Bénard and O. Hobert, unpublished data). The only zig genes that display a phenotype on their own are the previously identified zig-4 gene and the zig-3 gene that we describe here.
The zig-3 mutant phenotype corroborates key features of neuronal maintenance factors: (a) their absence causes no obvious developmental defects; (b) their activity is required during a critical period in the first larval stage; (c) they are not needed if locomotion of the animal is suppressed.
The function of zig-3 appears intimately tied to that of the zig-4 maintenance factor. Both genes are coexpressed by the same cell (the PVT neuron); their null mutant phenotypes are indistinguishable and not mutually enhanceable. One possible interpretation of the linked nature of these two factors is that they act as heterodimers. A dimeric configuration has been observed for many other Ig domain proteins, particularly of the cell surface type in the immune system, historically the most intensively studied Ig domain proteins (Barclay et al. 1997). An obligate heterodimer model is also consistent with our observation that one protein cannot substitute for the other, even in an overexpression scenario. However, the function of ZIG-3 and ZIG-4 may not need to be as tightly molecularly linked; it is also possible that ZIG-3 and ZIG-4 affect one common pathway at distinct steps.
ZIG-3 and ZIG-4 may act as signaling proteins that activate receptor system(s). Alternatively, ZIG-3 and ZIG-4 may be part of a multimeric adhesion complex required to hold axons together in a fascicle. They may provide adhesive activity within such a complex or may alter the adhesive ability of other molecules.
The analysis of zig-3 and zig-4 mutants, as well as laser ablation studies, reveal a remarkable complexity of the mechanisms that hold the ventral nerve cord together. The pushing of axons through mechanical stress is clearly a necessary component for the induction of the flip-over event, as flip-overs (induced by loss of maintenance factors or PVT ablation) can be suppressed through immobilizing animals. However, mechanical force is clearly not sufficient to push axons around. First, vigorous movement of animals is already evident in the threefold embryo, yet axon flip-overs observed upon loss of PVT or maintenance factors only occur in the first larval stage. Second, movement does not result in flip-overs if the contralateral homolog of an axon is removed. One could argue that in such a case flip-over may in fact occur, but that a flipped axon requires its contralateral homolog to stabilize its position after it has flipped over the midline, perhaps via homophilic interaction. In the absence of such stabilization, a flipped axon may simply not remain in the opposite fascicle and be pushed back into its original fascicle. Such “pushing back” may occur passively by an increase in size of the midline structure (“hypodermal ridge”) occurring during larval stages. While an attractive model, the laser ablation of another class of ventral cord neurons, the PVP neurons complicates this model. The suppression of the left PVQ axon flip-over into the right fascicle also occurs when the PVP axon that is in the left ventral nerve cord is ablated. This cannot be a result of lack of stabilization of the flipped axons in the opposite cord, but rather suggests that the left cord PVP axon somehow provides a latent impetus for the left PVQ axon to move away into the opposite fascicle. Even though we find it hard to attach a molecular mechanism to these findings, the most conservative explanation is that there appears to be a tight balance of latent attractive and repulsive events in the ventral nerve cord. Loss of PVT-mediated axon maintenance exposes axons to such latent attractive and repulsive forces, with axons being pushed and pulled over the midline. It will require the identification of more axon maintenance factors, and specifically a detailed analysis of their molecular interactions to more comprehensively understand these phenotypes.
We note that vertebrate nervous systems have to deal with similar challenges as the worm nervous system in that axon fascicles have to withstand mechanical stress from a variety of distinct sources (Bénard and Hobert 2009). Moreover, a plethora of as yet uncharacterized small Ig domain proteins are expressed in the vertebrate brain as well (http://www.brain-map.org/) (Barclay et al. 1997). If some of those molecules are indeed involved in maintaining nervous system structure, they may be excellent candidates to be involved in neuronal pathologies, particularly adult-onset ones that may reflect a failure to maintain the specific architectural features of a nervous system.
Acknowledgments
We thank Q. Chen for expert technical assistance, the Caenorhabditis Genetics Center and particularly the Caenorhabditis elegans knockout consortia in Oklahoma, Vancouver, and Tokyo, under the leadership of R. Barstead, D. Moerman, and S. Mitani, respectively, for providing knockout strains; L. Segalat for providing the transposon insertion strain; and many colleagues in the worm community for providing reporter transgenes. This work was funded in part by the Muscular Dystrophy Association, the Boehringer Ingelheim Fond (to T.B.), the Natural Sciences and Engineering Research Council of Canada, and Canadian Institute of Health Research (to C.B.) and the Howard Hughes Medical Institute.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.107441/DC1.
References
- Aurelio, O., D. H. Hall and O. Hobert, 2002. Immunoglobulin-domain proteins required for maintenance of ventral nerve cord organization. Science 295 686–690. [DOI] [PubMed] [Google Scholar]
- Aurelio, O., T. Boulin and O. Hobert, 2003. Identification of spatial and temporal cues that regulate postembryonic expression of axon maintenance factors in the C. elegans ventral nerve cord. Development 130 599–610. [DOI] [PubMed] [Google Scholar]
- Barclay, A. N., M. H. Brown, S. K. A. Law, A. J. McKnight, M. G. Tomlinson et al., 1997. The Leucocyte Antigen Facts Book. Academic Press, London.
- Bargmann, C. I., and L. Avery, 1995. Laser killing of cells in Caenorhabditis elegans. Methods Cell Biol. 48 225–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bénard, C., and O. Hobert, 2009. Looking beyond development: maintaining nervous system architecture. Curr. Top. Dev. Biol. 87 175–194. [DOI] [PubMed] [Google Scholar]
- Bénard, C. Y., A. Boyanov, D. H. Hall and O. Hobert, 2006. DIG-1, a novel giant protein, non-autonomously mediates maintenance of nervous system architecture. Development 133 3329–3340. [DOI] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bülow, H. E., T. Boulin and O. Hobert, 2004. Differential functions of the C. elegans FGF receptor in axon outgrowth and maintenance of axon position. Neuron 42 367–374. [DOI] [PubMed] [Google Scholar]
- Clark, S. G., and C. Chiu, 2003. C. elegans ZAG-1, a Zn-finger-homeodomain protein, regulates axonal development and neuronal differentiation. Development 130 3781–3794. [DOI] [PubMed] [Google Scholar]
- Fay, D., and A. Bender, 2008. SNPs: introduction and two-point mapping. WormBook September 25 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilleard, J. S., J. D. Barry and I. L. Johnstone, 1997. cis regulatory requirements for hypodermal cell-specific expression of the Caenorhabditis elegans cuticle collagen gene dpy-7. Mol. Cell. Biol. 17 2301–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert, O., 2002. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques 32 728–730. [DOI] [PubMed] [Google Scholar]
- Hutter, H., 2003. Extracellular cues and pioneers act together to guide axons in the ventral cord of C. elegans. Development 130 5307–5318. [DOI] [PubMed] [Google Scholar]
- Jin, Y., E. Jorgensen, E. Hartwieg and H. R. Horvitz, 1999. The Caenorhabditis elegans gene unc-25 encodes glutamic acid decarboxylase and is required for synaptic transmission but not synaptic development. J. Neurosci. 19 539–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntire, S. L., R. J. Reimer, K. Schuske, R. H. Edwards and E. M. Jorgensen, 1997. Identification and characterization of the vesicular GABA transporter. Nature 389 870–876. [DOI] [PubMed] [Google Scholar]
- Much, J. W., D. J. Slade, K. Klampert, G. Garriga and B. Wightman, 2000. The fax-1 nuclear hormone receptor regulates axon pathfinding and neurotransmitter expression. Development 127 703–712. [DOI] [PubMed] [Google Scholar]
- Notredame, C., D. G. Higgins and J. Heringa, 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302 205–217. [DOI] [PubMed] [Google Scholar]
- Ogura, K., M. Shirakawa, T. M. Barnes, S. Hekimi and Y. Ohshima, 1997. The UNC-14 protein required for axonal elongation and guidance in Caenorhabditis elegans interacts with the serine/threonine kinase UNC-51. Genes Dev. 11 1801–1811. [DOI] [PubMed] [Google Scholar]
- Okkema, P. G., S. W. Harrison, V. Plunger, A. Aryana and A. Fire, 1993. Sequence requirements for myosin gene expression and regulation in Caenorhabditis elegans. Genetics 135 385–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz, C. O., J. F. Etchberger, S. L. Posy, C. Frokjaer-Jensen, S. Lockery et al., 2006. Searching for neuronal left/right asymmetry: genomewide analysis of nematode receptor-type guanylyl cyclases. Genetics 173 131–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pocock, R., C. Y. Bénard, L. Shapiro and O. Hobert, 2008. Functional dissection of the C. elegans cell adhesion molecule SAX-7, a homologue of human L1. Mol. Cell. Neurosci. 37 56–68. [DOI] [PubMed] [Google Scholar]
- Rougon, G., and O. Hobert, 2003. New insights into the diversity and function of neuronal immunoglobulin superfamily molecules. Annu. Rev. Neurosci. 26 207–238. [DOI] [PubMed] [Google Scholar]
- Sarafi-Reinach, T. R., T. Melkman, O. Hobert and P. Sengupta, 2001. The lin-11 LIM homeobox gene specifies olfactory and chemosensory neuron fates in C. elegans. Development 128 3269–3281. [DOI] [PubMed] [Google Scholar]
- Sasakura, H., H. Inada, A. Kuhara, E. Fusaoka, D. Takemoto et al., 2005. Maintenance of neuronal positions in organized ganglia by SAX-7, a Caenorhabditis elegans homologue of L1. EMBO J. 24 1477–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., J. Kweon, S. Larson and L. Chen, 2005. A role for the C. elegans L1CAM homologue lad-1/sax-7 in maintaining tissue attachment. Dev. Biol. 284 273–291. [DOI] [PubMed] [Google Scholar]
- White, J. G., E. Southgate, J. N. Thomson and S. Brenner, 1986. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R Soc. Lond. B Biol. Sci. 314 1–340. [DOI] [PubMed] [Google Scholar]
- Zallen, J. A., S. A. Kirch and C. I. Bargmann, 1999. Genes required for axon pathfinding and extension in the C. elegans nerve ring. Development 126 3679–3692. [DOI] [PubMed] [Google Scholar]