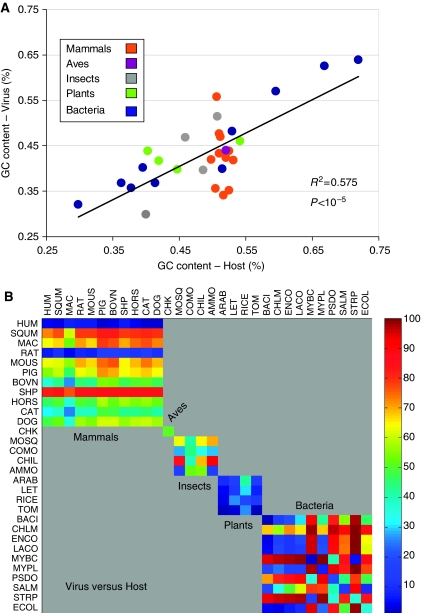
Figure 6.
Similarity in GC content and codon usage between pairs of viruses and hosts. The GC content from the proteomes of all viruses and their hosts was compiled. (A) Analysis of the GC content correlation between the hosts (x-axis) and viruses (F-test for linear regression), color coded by their taxonomical grouping to mammals, aves, insects, plants, and bacteria (according to Table I). (B) Codon usage distance matrix for all pairs of hosts and viruses is shown. Color code is according to the ranking of all 900 values as calculated from all pairs of 30 viruses and 30 unique hosts. The matrix is organized by groups according to Table I. L2 distance matrix for all 30 viruses (y-axis) and 30 unique hosts (x-axis). The analysis is based on the complete proteomes of the mapped viruses. The sub-matrices indicate the partition to groups of mammals (1–11), aves (12), insects (13–16), plants (17–20), and bacteria (21–30). Note the strong resemblance in human and rat viruses relative to all other mammals and the resemblance among all viruses infecting plants. For data of the complete matrix, see Supplementary information S2.