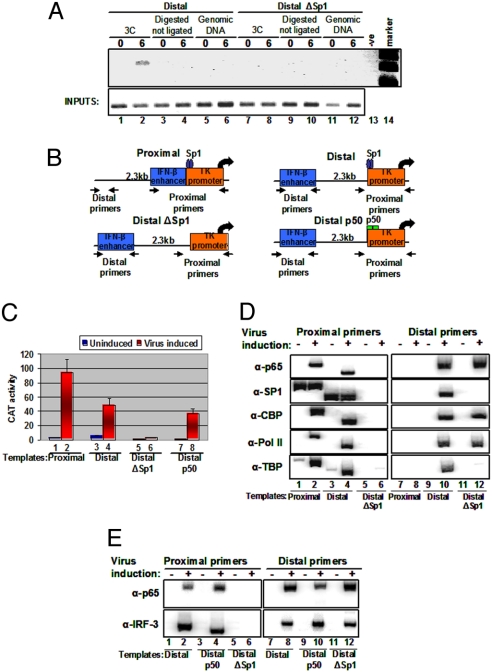
Fig. 2.
DNA looping mediates the interaction between a remote enhancer and a promoter. (A) Shown is a 3C experiment depicting the PCR products using primers specific for the enhancer and the promoter as seen in Fig. S1. PCR was performed on NlaIII-digested chromatin derived from HeLa cells mock or virus infected for 6 h harboring the Distal (lanes 1 and 2) or the DistalΔSp1 (lanes 7 and 8) plasmids. Genomic DNA (lanes 5, 6, 11, and 12) and cross-linked digested but not ligated chromatin (lanes 3, 4, 9, and 10) derived from mock- or virus-infected (6 h) cells were used as controls. Lane 13 is a negative PCR control, and lane 14 is the size marker. (B) Schematic representation of the CAT constructs used to determine mechanisms of enhancer function. The arrows indicate the position of the primers used in the PCR reactions with immunoprecipitated DNA. The wild-type TK promoter contains an Sp1 site (Proximal and Distal constructs), whereas in the DistalΔSp1 construct the Sp1 site has been mutated. In the Distal p50 construct, the Sp1 site was replaced by a consensus p50 homodimer site. (C) Stable HeLa cells bearing the indicated CAT reporter plasmids were mock or virus infected for 12 h before being harvested; then CAT activity was determined. The error bars indicate SD. (D) Cross-linked chromatin prepared from mock- or virus-infected (6 h) HeLa cells stably transfected with the indicated CAT constructs was immunoprecipitated with the indicated antibodies. The precipitated DNA was subjected to PCR analysis using 32P-dCTP and plasmid-specific primers. (E) The process is as described in (D), except that p65 and IRF-3 antibodies were used, and the Distal p50 construct instead of the proximal construct was included in the experiment.