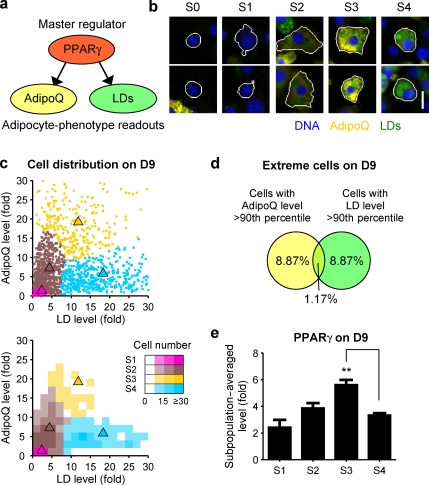
Figure 2.
Automated clustering identified differentiating 3T3-L1 subpopulations with distinct phenotypes. (a) AdipoQ and LDs were chosen as downstream readouts of PPARγ. (b) Differentiating 3T3-L1 cells from D6–12 (Fig. S1 c) were computationally pooled and clustered based on their AdipoQ and LD levels. Four subpopulations were identified (S1–4). The subpopulation of quiescent cells (S0) was excluded before clustering. Immunofluorescence images of two cells near the centroid of each subpopulation are shown. White lines indicate cell segmentation boundaries. (c) Scatter plot and 2D histogram showing the distribution of 3T3-L1 cells on D9. Triangles indicate centroids for S1–4 in the original subpopulation model. (d) Venn diagram showing the overlap of 3T3-L1 cells with high AdipoQ or LD levels. (e) Subpopulation-averaged levels of PPARγ. Error bars indicate SEM (n = 3); **, P < 0.01 by two-tailed paired t test. Bar, 20 µm.