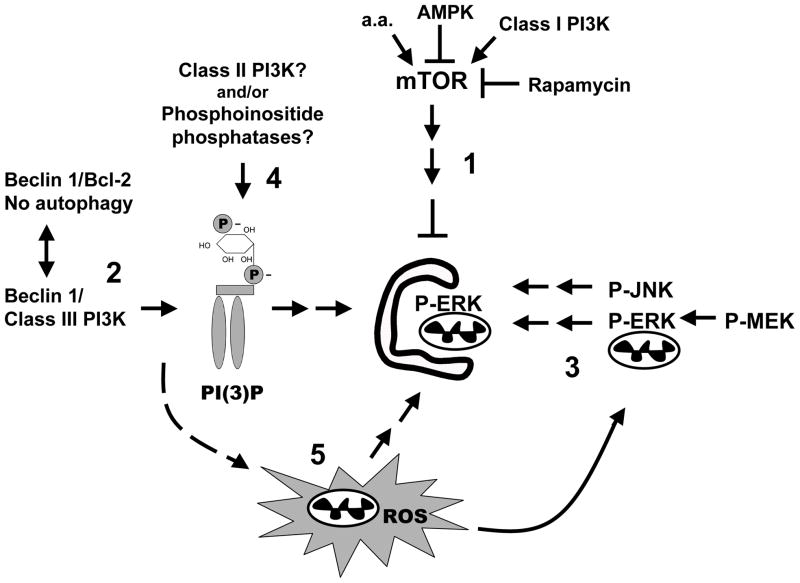
Fig. 1. Schematic diagram of potential regulatory pathways for autophagy/mitophagy.
The most studied pathways of autophagy regulation, including mTOR-mediated suppression of autophagy, which is reversed by amino acid deprivation, rapamycin, or AMP-kinase (1), and the Beclin 1-class III PI3K pathways (2) are shown. In addition, there is cell type-dependent stimulation of autophagy by ERK and JNK signaling (3), although kinase targets remain unidentified. Alternative mechanisms for generating PI(3)P exist in mammalian cells, including wortmannin-resistant class II PI3Ks and phosphoinositide phosphatases (4, see text). Recent studies also suggest a role for ROS acting downstream of Beclin 1 to reduce cleavage of LC3 from preautophagosomal membranes (5). The signals involved in generating mitochondrial ROS during starvation remain undefined, but ROS are involved in mitochondrial activation of ERK. It is unknown if ERK acts directly to phosphorylate mitochondrial targets or if its effects on autophagy reflect other cytoplasmic sites of action.