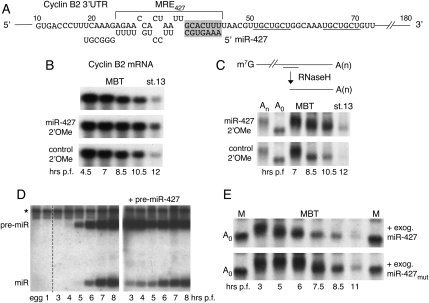
FIGURE 2.
miR-427-dependent destabilization and deadenylation of cyclin B2 mRNA. (A) Nucleotide sequences in the cyclin B2 3′ UTR showing the predicted seed-match (shaded box) for miR-427 within a potential MRE427, and potential miR-16 seed-matches (underlined). (B) Stabilization of cyclin B2 mRNA upon inactivation of miR-427. Northern blot analyses of endogenous cyclin B2 mRNA in early embryos that were untreated (top) or injected at the one-cell stage with 2′-OMe-antisense oligonucleotides against miR-427 (middle) or a let-7 control (bottom). (C) Impaired deadenylation of endogenous cyclin B2 mRNA upon inactivation of miR-427. (Top) Schematic of RNase H assay for determination of changes in poly(A) tail length, using a deoxyoligonucleotide that targets sequences ∼500 nt from the 3′ end of the mRNA. (Lower panels) Northern blot analyses of the 3′-terminal fragments of cyclin B2 mRNA generated by digestion with RNase H, showing the kinetics of deadenylation in the absence (top) or presence (bottom) of functional miR-427. Lanes An and A0 show size markers generated by digestion of cyclin B2 mRNA of pre-MBT embryos with RNase H plus the deoxy-oligonucleotide, in the absence (An) or presence (A0) of oligo-dT15. (D) Northern blot analyses of pre-miR-427 and miR-427 in normal embryos (left) and in embryos injected with exogenous pre-miR-427 (right); a similar pattern of miRNA accumulation was seen when pre-miR-427mut was injected. Analyses of RNAs from unfertilized eggs and one-cell (1 h p.f.) embryos (left panel) show the lack of pre-miR- and miR-427 in those cells. (E) Accelerated deadenylation of endogenous cyclin B2 mRNA upon premature expression of miR-427, but not miR-427mut. Exogenous pre-miR-427 or pre-miR-427mut was injected into one-cell embryos, and RNase H digested RNAs were analyzed as in C.