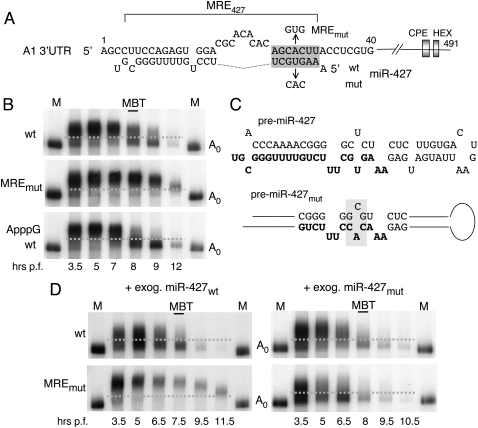
FIGURE 5.
MRE•miRNA base pairing in the deadenylation of cyclin A1 3′ UTR RNA. (A) Nucleotide sequences in the cyclin A1 3′ UTRs (top strand) showing the predicted seed-match (shaded box) and upstream 3′ UTR sequences that allow for functional pairing with miR-427 (bottom strand; cf. Fig. 6). The inactivating mutation of the MRE (MREmut) and the compensatory change in the seed sequence miR-427 (mut) are indicated. (B) Deadenylation of 32P-labeled A1 3′ UTR reporter RNAs lacking a coding region and containing either a wild-type (wt) or mutant MRE (MREmut), or an ApppG- cap, to ensure lack of translation. (C) Sequences of wild-type and mutant forms of pre-miR-427. Nucleotide changes in pre-miRmut (shaded) were designed to encode miR-427mut with an altered seed sequence; (bold) mature miRNA sequences. (D) Rescue of MREmut by the compensatory mutation of the miR-427 seed sequence. Kinetics of polyadenylation and deadenylation of cyclin A1 3′ UTR reporter RNAs with wild-type (top panels) or mutant (bottom panels) seed-matches; the reporters were co-injected with exogenous pre-miRNAs generating either wild-type (left panels) or mutant (right panels) miR-427.