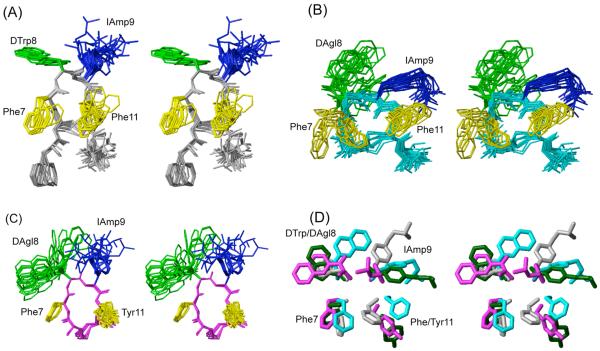
Figure 2.
3D NMR structures of analogues (A) H-DPhe-c[Cys-Phe-DTrp-IAmp-Phe-Cys]-Thr-NH2 (16), (B) H-c[Cys-Phe-DAgl(NMe,2naphthoyl)-IAmp-Phe-Cys]-Thr-NH2 (23) and (C) H-c[Cys-Phe-DAgl(NMe,2naphthoyl)-IAmp-Tyr-Cys]-NH2 (27). For each analogue, 20 energy-minimized conformers with the lowest target function are used to represent the 3D NMR structure. The bundle is obtained by overlapping the Cα atoms of all the residues. The backbone and the side chains are displayed including the disulphide bridge. The amino acid side chains that are proposed to be involved in sst1 binding are highlighted: light green, DTrp/DAgl at position 8; blue, IAmp at position 9; yellow, Phe at positions 7 and 11. In (D) the side chains of sst1 binding analogue 5 (in dark green) are superimposed on the structure of 16 (in grey), 23 (in cyan) and 27 (in magenta). It should be noted that the aromatic side chains at position 7 and 11 of analogue 5 are in the other side of the peptide backbone compared to 16, 23 and 27.