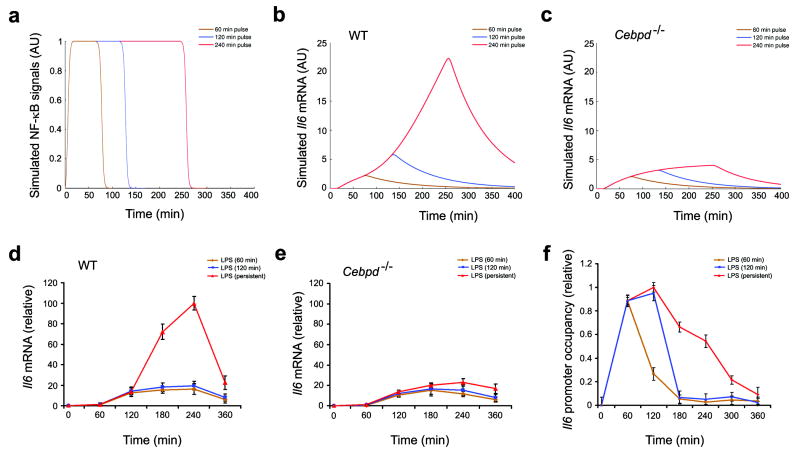
Figure 3. Computational simulations of the transcriptional response of Il6 to TLR4 signals of varying duration reveal a threshold effect.
a, Computational simulation of transient and persistent NF-κB signals. To simulate LPS pulsing, the NFκB signal was computationally varied over time. NF-κB signals of the same amplitude but different duration are shown.
b,c Outputs of computationally simulated Il6 transcriptional response to transient LPS signals in WT (b) and Cebpd-/- (c) macrophages.
d,e Macrophages from wild-type (d) and Cebpd-/- (e) mice were stimulated for 1 or 2 h or persistently with 10ng/ml LPS. Cells were harvested at the indicated time points and Il6 mRNA was measured by quantitative real-time RT-PCR. Data points represent the average of three independent experimental values and error bars indicate ± standard error.
f, Macrophages from wild-type mice were stimulated for 1 h or 2 h or persistently with 10 ng/ml LPS. Cells were harvested at the indicated times and processed for ChIP assays as in Fig. 1c. Presence of immunoprecipitated Rel, on the Il6 promoter was measured by quantitative real-time RT-PCR. Transcription factor binding was normalized to the amount of PCR product loaded. Data points represent the average of three independent experimental values and error bars indicate ± standard error.