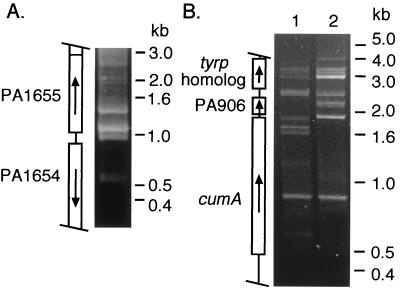
Figure 4.
Genetic footprinting at three targeted regions of the chromosome. Plasmid libraries of in vitro generated transposon insertions in pSW1654–55 and pSW906 were mobilized from E. coli to SWSce. After selection with the appropriate antibiotics, mutant pools were used for PCR analysis. Locations of the transposon insertions are aligned with the corresponding gene position. Arrows indicate direction of transcription. (A) Genetic footprinting at the PA1655 locus and part of the PA1654 gene. Colony PCR was performed with primers MarOUT and MERPA1654 located near the 3′ end of PA1654 on mutant pools. (B) Genetic footprinting at the PA906 region. PCR analysis was performed with primers MarOUT and MER906G1 located ≈2.0-kb from the 5′ end of the putative start site for PA906 (lane 1) on genomic DNA isolated from mutant pools. The inferred putative start site is based on the sequence of the PA906 homolog, cumB from Pseudomonas putida (26). The same pSW906 mutagenized plasmid library was introduced into strain SWSce2.6 complemented with a wild-type copy of PA906. Genomic DNA from mutant pools was used as template in PCR with primers MarOUT and MER906G1 (lane 2).