Abstract
Polyethylenimine has been used widely in transient gene expression with mammalian cells. To further understand its mediation of gene transfer, the transfection of HEK 293-F cells with dynamically prepared PEI/DNA complexes was studied with the help of fluorescent labeling. The efficiency of complex endocytosis/phagocytosis was found to correlate with the average sizes of complexes applied and complexes greater than 1 μm in diameter were likely excluded by the cells. Coupled with complex growth in size, the degree of association between PEI and DNA increased with the time of complex formation in the presence of competing ions. The blocking of transcription by complex formation necessitated complex dissociation in the nuclear environment for transcription to happen. Intracellularly, the fates of PEI complexed DNA therefore may be mostly determined by the degree of association. Results also suggested that the uptake of PEI/DNA complexes and subsequent protein expression were independent of the cell cycle stages of HEK 293-F cells.
Keywords: Cell cycle, Flow cytometry, HEK 293, Polyethylenimine, Particle size, Transient gene expression
Introduction
Polyethylenimine (PEI)-mediated transient gene expression processes are expected to play an increasingly important role in the development of protein therapeutics within the scope of biopharmaceutical applications (Baldi et al. 2007). Serving primarily as an alternative tool for recombinant protein preparation, however, these processes have generally been found to be relatively less productive compared to those of well-established stable expression systems such as CHO and NS0 cells (Girard et al. 2002; Wurm 2004; Baldi et al. 2005). While moderate improvements could be made by optimizing the procedure of transfection or adopting operational strategies of high cell density and fed-batch cultures (Girard et al. 2001; Sun et al. 2006, 2008; Han et al. 2007), a recent sophisticated implementation involving the rational design of expression vectors and use of cell cycle regulators has led to a recombinant antibody titer of over 1 g L−1 in serum-free media (Backliwal et al. 2008).
In spite of the current availability of various types of PEIs, recent studies have recognized the associations of their transfection efficiency and cytotoxicity with their structural complexities and molecular weights (Kunath et al. 2003; Derouazi et al. 2004; Lungwitz et al. 2005; Werth et al. 2006). As a result, a linear 25 kDa PEI in particular, with which transfection efficiencies of 40–60% were reportedly achieved, is now commonly chosen to transfect mammalian cells such as HEK 293 and CHO cells grown in suspension cultures (Schlaeger and Christensen 1999; Derouazi et al. 2004; Han et al. 2007).
In the presence of PEIs, DNA molecules are condensed and compacted into PEI/DNA complex particulates, acquiring the capability to interact with electrostatically negative moieties such as heparin sulfate proteoglycans on the surfaces of cells for endocytosis/phagocytosis (Godbey et al. 1999; Kopatz et al. 2004). Intracellularly, endocytosed complexes are subsequently transported to the subcellular membrane structure endosomes and/or lysosomes. The currently accepted proton sponge effect explains their escape from these organelles by the swelling of endosomes/lysosomes and prevention of enzymatic degradation of DNA molecules (Boussif et al. 1995). Since DNA molecules microinjected into the cytoplasm in their naked forms were unable to accomplish their nuclear entry efficiently, they are supposed to remain complexed afterwards in order to enter the nuclei of the cells transfected (Capecchi 1980; Forbes 1992; Godbey et al. 1999). Ultimately, the nuclear status of DNA molecules may determine their transcriptability for the expression of protein products. From the formation of PEI/DNA complexes to the expression of the transgene, the polycationic nature of PEIs apparently plays a critical role in all these physiochemical events.
In this study, the endocytosis/phagocytosis of PEI/pEGFP-N1 complexes dynamically prepared and the expression of green florescent protein (GFP) were investigated with suspension grown 293-F cells to identify the qualities of PEI/DNA complexes which may be critical to their uptake and subsequent gene expression. Microcaloric and surface plasmon resonance assessments were made to characterize the interaction patterns between PEI and DNA molecules under different binding conditions. The examination of the intracellular statuses of PEI and DNA helped to identify their possible relationships with their nuclear entry and gene expression. In vitro transcription and gel retardation assays were performed to reveal the relationship between complex formation and gene transcription. The relationship of cell cycle with the endocytosis/phagocytosis and expression of complexed pEGFP-N1 was also investigated. The authors believe these findings could be valuable for the development of novel implementations to the PEI-mediated gene expression processes.
Materials and methods
Covalent labeling of DNA and PEI
Circular 4.7 kbp plasmid DNA pEGFP-N1 (Clontech) was covalently labeled with Cy3 or Cy5 at a frequency of 1 fluorophore per 30 bp in average with a Label IT Kit (Mirus) and purified with a DNA Midi Purification Kit (TianGen), according to manufacturer’s instructions.
Linear PEI (25 kDa, Polysciences) was covalently labeled by suspending one vial of Cy5 mono-reactive dye (Amersham) in 1 mL of 2 mg mL−1 PEI dissolved in 0.1 M NaHCO3 (pH 9.3) for 1 h at room temperature with intermittent mixing at an interval of 10 min. A 10 × 110 mm Sephacryl S-200 HR (Amersham) column was used to eliminate free Cy5. Fractions from 4.5 to 9.5 min were collected and pooled when the column was eluted with 150 mM NaCl at 1 mL min−1. The concentration of PEI was quantified by spectrophotometry analysis according to the method previously described (Ungaro et al. 2003). The concentration of Cy5 was estimated with a molar extinction coefficient of 250,000 M−1 cm−1 at 650 nm. Based on the molar concentrations of PEI and Cy5 calculated, the labeling frequency of PEI was estimated to be 1 fluorophore per 4 free amidoes in average.
PEI/DNA complexes preparation and cell transfection
PEI/pEGFP-N1 complexes were prepared by mixing equal volumes of 120 μg mL−1 PEI and 20 μg mL−1 pEGFP-N1 in 150 mM NaCl (pH 6.8) as described previously (Han et al. 2007). N/P ratio, which is the molar ratio of the cationic PEI nitrogen to the anionic DNA phosphate, was estimated to be ~46 according to the amounts of PEI and DNA applied and their structural information. Samples of complexes formed at varying time points were taken for laser-light scattering size analysis with a Malvern Zetasizer Nano S (Malvern).
Suspension culture adapted 293-F cells (Invitrogen), grown in protein-free CD 293 medium (Invitrogen) in shaker flasks at 37 °C in a 5% CO2 atmosphere, were routinely passaged between 2.0 × 105 and 3.0 × 106 cells mL−1. 293-F cells were transfected with an optimized procedure as reported previously unless specified (Han et al. 2007). Briefly, exponentially growing 293-F cells were harvested by centrifugation and resuspended in fresh low calcium (100 μM) RPMI 1640 to the density of 1.0 × 106 cells mL−1 before being seeded in 6-well plates (Corning). PEI/pEGFP-N1 complex preparations of varying formation time as above were added to cultures. The final concentrations of PEI and pEGFP-N1 in transfection cultures were 6 and 1 μg mL−1, respectively. An equal volume of CD 293 medium was then added to each culture 4 h later for post-transfection cultures.
Isothermal titration calorimetry (ITC) and surface plasmon resonance (SPR) of PEI and DNA interaction
For ITC investigation on the interaction between DNA and PEI, 270 μL of PEI (3 mg mL−1 in 150 mM NaCl, 10 mM HEPES, pH 7.0) was injected into 1,468 μL of DNA (0.725 mg mL−1 DNA in the same) on an ITC-100 (MicroCal, LLC). Both concentrations were elevated to compromise the instrumental sensitivity of measurement. However, the ratio of PEI to DNA was unable to remain the same as that for transfection because of the limitation of PEI solubility in the buffer used.
SPR measurements were performed on Biacore 3000 (Biacore AB). The gold sensor chip was modified with 11-mercaptoundecanol, carboxylated dextran and streptavidin (Löfås and Johnsson 1990). Biotin-labeled DNA (1.1 kbp), generated by PCR amplification with biotinylated oligonucleotides as the primers and pEGFP-N1 as the template, was immobilized on one of the flow cell through biotin-streptavidin interaction. The other flow cell without the biotin-labeled DNA was used as a reference. All measurements were performed at 25 °C using a running buffer (0.01% Tween-20, 150 mM NaCl, 10 mM HEPES, pH 7.0) at a constant flow rate of 10 μL min−1. Injections of 10, 30, 50 and 100 μL of PEI (12.5 μg mL−1 in 150 mM NaCl, 10 mM HEPES, pH 7.0) were made to the system, corresponding to an interaction time of 1, 3, 5 and 10 min, respectively. The dissociation phases of PEI/DNA complexes were recorded. The sensor chips were regenerated with 50 μL of 2 M NaCl and re-equilibrated with running buffer before next injection. The effects of pH (10 mM HEPES, 150 mM NaCl) and NaCl concentration (10 mM HEPES, pH 7.0) were investigated on the interaction between immobilized DNA and free PEI for 5 min, and dissociations of PEI from immobilized DNA in the running buffer afterwards.
Flow cytometry analysis of cell cycle
Before nuclear staining with propidium iodide (PI), the cells were fixed and permeabilized (Chu et al. 1999). Briefly, 1–2 × 106 293-F cells, twice washed with PBS (containing 300 mg L−1 heparin) and suspended in 0.5 mL of ice cold PBS, were first mixed with 0.5 mL of 2% (v/v) paraformaldehyde in PBS and incubated for 1 h at 4 °C. The cells were then permeabilized by an overnight treatment in 1 mL of 70% ethanol at 4 °C. Chromosomal staining of cells suspended in PBS was executed by the addition of PI together with Ribonuclease A (Sigma) at final concentrations of 20 μg mL−1 and 200 μg mL−1, respectively, and an incubation in the dark for 30 min at 37 °C. A minimum of 10,000 events of viable cells only was collected per sample for analyses on a FACSAria (BD Biosciences) equipped with CellQuest software. Analyses of the multivariate data and cell cycle of DNA histograms were performed with FCS Express software and MultiCycle software (De Novo software), respectively. It should be noted that samples of cells were washed with heparin containing PBS before assessments to eliminate any complexes on the surfaces of the cells.
Nucleus isolation
The nuclei of 239-F cells were isolated hypoosmotically (Ausubel et al. 2002). Briefly, cells washed in ice cold PBS were resuspended in ice cold NP-40 lysis buffer, a 0.5% (v/v) NP-40 solution containing 10 mM NaCl, 3 mM MgCl2 and 10 mM Tris–HCl (pH 7.4). After 5 min of incubation on ice, lysates were microscopically examined to ensure a completion of lysis and liberation of nuclei from cytoplasmic materials. With the help of centrifugation, the nuclear pellets were further washed twice in the lysis buffer and finally resuspended in suspension buffer (3 mM CaCl2, 2 mM MgCl2, 10 mM Tris–HCl, pH 7.4).
Confocal laser scanning microscopy (CLSM)
To examine the intracellular events of dual-labeled complexes (Cy5-PEI/Cy3-pEGFP-N1) after endocytosis, samples of cells transfected were washed with heparin containing PBS (300 mg L−1) and resuspended in PBS before being aliquoted onto polylysine-coated slides (Ausubel et al. 2002). After an incubation of 10 min at 37 °C, cells attached were further treated with 4% paraformaldehyde for 10 min. Specimens were sealed with an anti-fading reagent after nuclear staining with Hoechst 33342 (2 μg mL−1 in PBS). Images collected with a Zeiss LSM 510 META confocal laser scanning microscope were analyzed with LSM 5 software. Samples of cells were washed with heparin containing PBS before assessments for the same reason as above.
In vitro transcription and agarose gel retardation
To investigate the effect of complex formation on transcription, the activity of transcription of linear pGEMEX-1 (3.6 Kbp) containing T7 promoter region was measured using Riboprobe® in vitro Transcription System (Promega). For DNA levels of 10 and 20 μg mL−1, free or PEI-complexed pGEMEX-1 was incubated at 37 °C for 1 h with an equal volume of 2× transcription mixture, respectively. RNA levels of transcription were then analyzed on 1% agarose gel.
To determine the binding capability of PEI to DNA, the agarose gel retardation assay was performed. PEI/pEGFP-N1 complexes were prepared by mixing varying concentrations of PEI (0–120 μg mL−1) with an equal volume of pEGFP-N1 (20 μg mL−1). After an incubation of 30 min at room temperature, electrophoresis was carried out in 1% agarose gels containing EB (0.5 μg mL−1) at a potential of 90 V. Bands of DNA were visualized by UV transillumination.
Results
Dynamic PEI/DNA complex preparation and 293-F transfection
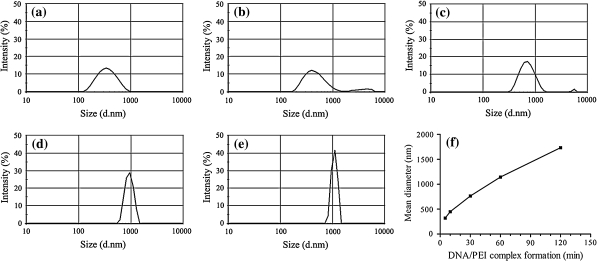
PEI/pEGFP-N1 complexes of different sizes were first prepared in a controlled manner with a previously optimized procedure (Han et al. 2007). Upon the mixing between the solutions of PEI and pEGFP-N1, the whole population of complexes observably experienced a dynamic shift of increasing in size temporally during the time period tested (Fig. 1). The average diameters of complexes formed were estimated to be 321, 449, 763, 1,143 and 1,734 nm, corresponding to a complex formation time of 5, 10, 30, 60 and 120 min, respectively.
Fig. 1.
Dynamics of PEI/pEGFP-N1 complex formation between equal volumes of 120 μg mL−1 PEI and 20 μg mL−1 pEGFP-N1 in 150 mM NaCl (N/P ratio 46). Particle sizing was performed on a Malvern Zetasizer Nano S. a 5 min; b 10 min; c 30 min; d 60 min; e 120 min. The average diameter of complexes plotted against complex formation time (f)
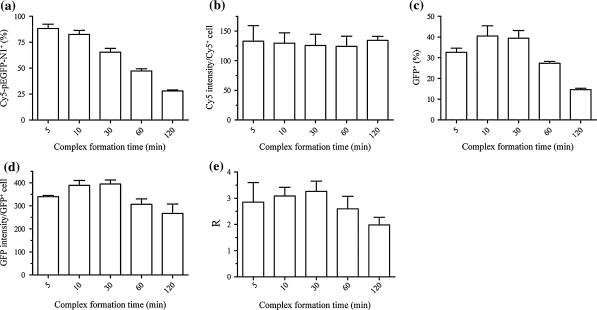
The above complex preparations were used to transfect 293-F cells. The tolerance of 293-F cells to the chosen level of PEI has been demonstrated previously concerning cytotoxicity and cell aggregation (Han et al. 2007). After a post-transfection culture of 20 h in CD 293, FACS assessment of intracellular Cy5 showed that the sizes of complexes affected the uptake of complexed Cy5-pEGFP-N1 adversely (Fig. 2a), suggesting an increasing difficulty in their cellular uptake or a size exclusion effect on the complexes as they became larger (Fig. 1). Interestingly, there existed no significant differences among the groups in the average level of intracellular Cy5-pEGFP-N1 in Cy5-pEGFP-N1 positive cells (Fig. 2b). The percentages of GFP positive cells in the total populations (transfection efficiencies) noticeably correlated with those of Cy5-pEGFP-N1 positive with an exception of the complex preparation of 5 min which was observed with the highest efficiency of endocytosis/phagocytosis (Fig. 2c). A similar trend was observed from the calculated average intensities of GFP in GFP positive cells (Fig. 2d). The expression of GFP could be enhanced if complex formation was extended from 5 min to 10–30 min; a further extension affected GFP expression negatively however (Fig. 2c, d). As a possible indicator of apparent intracellular activity of transcription and translation, the quotient of the average GFP intensities (Fig. 2d) to the average Cy5 intensities (Fig. 2b) demonstrated a close connection of pEGFP-N1 expression with the time of complex formation (Fig. 2e).
Fig. 2.
FACS analyses of 293-F cells transfected with PEI/Cy5-pEGFP-N1 complexes prepared in 150 mM NaCl (pH 6.8, N/P ratio 46) after transfection of 4 h and post-transfection culture of 20 h. a Percentages of Cy5-pEGFP-N1 positive cells in total populations; b average Cy5 intensities of Cy5-pEGFP-N1 positive cells; c percentages of GFP positive cells in total populations; d average GFP intensities of GFP positive cells; e ratios (R) of average GFP intensities of GFP positive cells to average Cy5 intensities of Cy5-pEGFP-N1 positive cells
Instrumental analyses of the interaction between immobilized DNA and free PEI
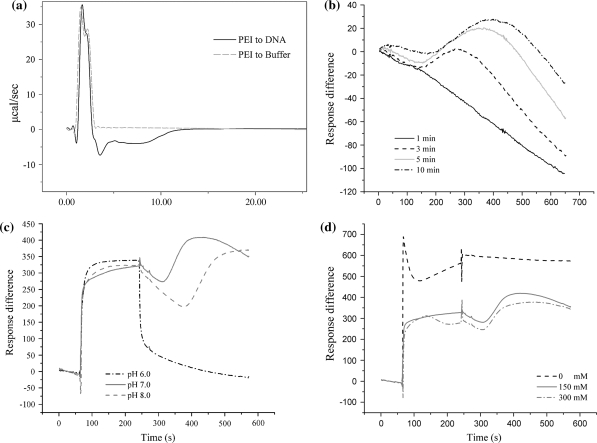
For the convenience of ITC measurement, elevated concentrations of DNA and PEI were employed in the experimental execution. Figure 3a clearly demonstrated a biphasic pattern upon the mixing of the two polymeric solutions, i.e., an early endothermic stage of mixing and a following exothermic stage of interaction between the two kinds of counterpart macromolecules in solution.
Fig. 3.
Instrumental analyses of the interaction between DNA and free PEI. a Isothermal titration calorimetry (ITC) measurement during complex formation; b dissociations of PEI from immobilized DNA; effects of pH (10 mM HEPES, 150 mM NaCl) (c) and NaCl concentration (10 mM HEPES, pH 7.0) (d) on the interaction between immobilized DNA and free PEI, and dissociations of PEI from immobilized DNA
In the SPR analysis, extending the time of binding between DNA and PEI clearly increased the difficulty for PEI to dissociate from the immobilized DNA as the direct result of their enhanced association in the presence of 150 mM NaCl (pH 7.0) (Fig. 3b). Within the range tested (6.0–8.0) for complex formation, pH did not make much difference in PEI binding but indeed affected PEI dissociation behavior significantly (Fig. 3c). Interestingly, the absence and presence of NaCl (150 and 300 mM) made significant differences in both the binding and the dissociation of PEI in the running buffer (Fig. 3d).
Cell cycle and PEI/DNA complex endocytosis
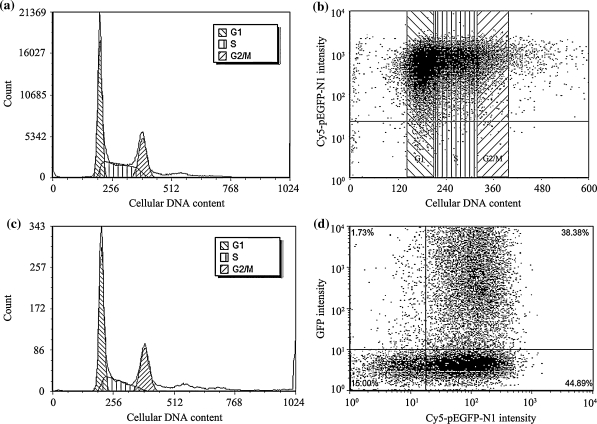
Approximately 43.6, 23.5 and 32.9% of the cell population were found in G1, G2/M and S phases after transfection for 4 h with PEI/Cy5-pEGFP-N1 prepared in 150 mM NaCl (N/P ratio 46, 10 min, mean diameter 449 nm) (Fig. 4a). Simultaneous FACS assessment of intracellular Cy5-pEGFP-N1 showed the vast majority of the cells intracellularly containing Cy5-pEGFP-N1, suggesting the cells were amendable to PEI-mediated gene transfection regardless of their phases in the cell cycle (Fig. 4b). Although GFP expression was detectable in only 1.8% of the total population right after transfection for 4 h only (Fig. 4c), its frequency distribution closely resembled that of the total population (Fig. 4a). After a post-transfection culture in CD 293 for 20 h, further analyses revealed no direct correlation between the levels of pEGFP-N1 endocytosed and GFP expression (Fig. 4d). Approximately half of the pEGFP-N1 positive cells failed to express GFP.
Fig. 4.
FACS analyses of 293-F cells after transfection with PEI/Cy5-pEGFP-N1 complexes (10 min preparation, N/P ratio 46) for 4 h without post-transfection culture (a–c) and with post-transfection culture of 20 h (d). a Cell cycle frequency distribution of the total population; b Cy5-pEGFP-N1 level in cells in different phases of cell cycle among the total population; c cell cycle frequency distribution of GFP positive cells in the total population; d dot plots of GFP intensity against Cy5 intensity displayed by all cell population. Non-transfected cells were used as controls
Confocal laser scanning microscopic observations
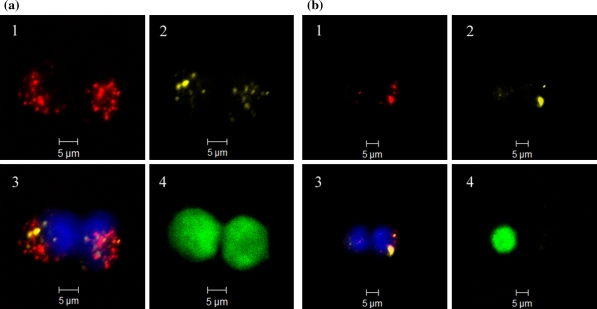
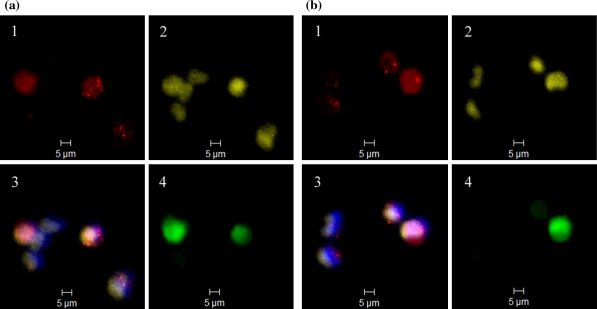
Transfection of 293-F with double fluorescently labeled complexes (N/P ratio 46, 10 min) enabled the examination of intracellular statuses of pEGFP-N1 (Cy3) and PEI (Cy5) under CLSM. After 14 h of post-transfection culture, PEI and pEGFP-N1 molecules were observed to co-exist dominantly as intracellular complex particulates (Fig. 5a). The complexes endocytosed were found to be mostly smaller than 1 μm in size. The expression of GFP seemed to be related to the presence of pEGFP-N1 in the form of minor particulates of complexes together with cloud-like dispersed form of pEGFP-N1 overlapping with the nuclear region. In spite of the fact that complexes still could be located in the proximities of the nuclei of two daughter cells after an extended post-transfection culture of 24 h, their persistence in the complexed form might lead to uneven distributions among daughter cells after mitoses (Fig. 5b). The presence of pEGFP-N1 in the dispersed form together with a minor aggregate enabled the expression of GFP within the daughter cell on the left, in contrast to the presence of larger complexes in the one on the right (Fig. 5b). Although the results of fluorescence were not quantitative, both pEGFP-N1 and PEI appeared to have undergone a transformation from the aggregated form to a mostly dispersed form at 48 h after transfection probably as the result of complex dissociation (Fig. 6). Different fluorescent patterns were observed among the cells for pEGFP-N1, PEI, nuclei and GFP. It is interesting that GFP expression seemingly required the co-existence of pEGFP-N1 and PEI since the cells containing dispersed pEGFP-N1 alone failed to express GFP in the absence of PEI (Fig. 6).
Fig. 5.
CLSM images of 293-F cells after post-transfection culture of 14 h (a) and 24 h (b) following transfection of 4 h with Cy5-PEI/Cy3-pEGFP-N1 (10 min, N/P ratio 46). (1) Cy5-PEI (red); (2) Cy3-pEGFP-N1 (yellow); (3) overlay of Cy5-PEI (red), Cy3-pEGFP-N1 (yellow) and Hoechst 33342 stained nuclei (blue); (4) intracellular GFP (green). (Color figure online)
Fig. 6.
CLSM images of 293-F cells after post-transfection culture of 48 h following transfection of 4 h with Cy5-PEI/Cy3-pEGFP-N1 (10 min, N/P ratio 46). Group a and b represent separate observations made respectively. (1) Cy5-PEI (red); (2) Cy3-pEGFP-N1 (yellow); (3) overlay of Cy5-PEI (red), Cy3-pEGFP-N1 (yellow) and Hoechst 33342 stained nuclei (blue); (4) intracellular GFP (green). (Color figure online)
In vitro transcription and PEI-DNA association
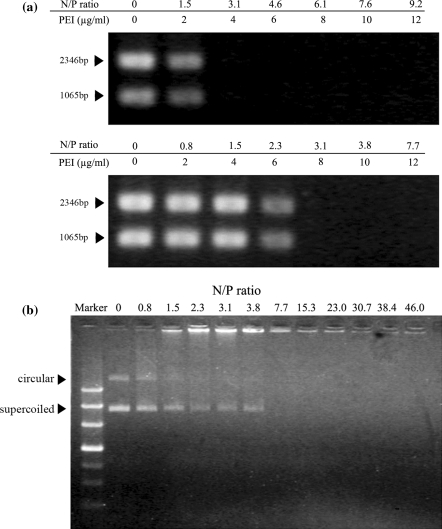
Since the transfer of transgenes across the nuclear membranes is a PEI aided process (Thomas and Klibanov 2003), the degree of association between PEI and plasmid DNA may have a direct impact on the transcriptability of a transgene in the nucleus. Transcription inhibition assays were performed on pGEMEX-1, free or PEI-complexed at different N/P ratios, to simulate the possible statuses of DNA molecules in the nuclei. For the selected levels of pGEMEX-1, 10 and 20 μg mL−1, increasingly partial inhibition of transcription activity was initially observed as the N/P ratio increased. Transcription activity switched off completely when the N/P ratio increased to a point below 3.1 in both cases (Fig. 7a). A test simulating the experimental conditions by incorporating pGEMEX-1 to the mixtures of PEI and the transcription reagent excluded the possibility of PEI interference to transcription activity (data not shown). These results clearly indicate the association of pGEMEX-1 with PEI adversely affected its accessibility for transcription.
Fig. 7.
In vitro transcription and gel retardation assays at varying N/P ratios. a PEI/pGEMEX-1 complexes of 0–9.2 N/P ratios were prepared after 30 min incubation in 150 mM NaCl before transcription mixture was added. Two RNA transcripts (1,065 and 2,346 bp) were expected according to the manufacturer’s instruction. Template concentration: top 10 μg mL−1; bottom 20 μg mL−1. Arrows indicate the positions of RNA transcribed. b PEI/pEGFP-N1 complexes were prepared by mixing 20 μg mL−1 pEGFP-N1 and varying concentrations of PEI for N/P ratios of 0–46 in 150 mM NaCl for 30 min. Arrows indicate the positions of uncomplexed DNA (circular and supercoiled)
In the estimation of the association between PEI and plasmid DNA with the agarose gel retardation assay, decreasing fractions of pEGFP-N1 remained free and migrated into the agarose gel as the N/P ratio increased initially, as indicated by two distinct fluorescent bands of super-coiled and circular DNA (Fig. 7b). The absence of DNA bands indicated the completion of pEGFP-N1 into complexes at the N/P ratios higher than 3.8. A conclusion drawn from the above results is that a full or partial dissociation of PEI/DNA complexes is necessary for transcription to initiate.
Discussion
Although 25 kDa linear PEI has been the most commonly used transfection reagent for transient gene expression in mammalian cells (Schlaeger and Christensen 1999; Derouazi et al. 2004; Baldi et al. 2005, 2007; Han et al. 2007; Backliwal et al. 2008), scientific questions remain regarding its association with the transgenes in the mediation of gene transfer from the extracellular to the nuclear environments and the initiation of gene transcription. It might be true that the fates of transgenes carried by PEI/DNA complexes are largely defined in the phase of complex formation. Complex preparation is therefore one of the critical steps to maximize the outcomes of PEI-mediated protein expression processes.
Polyanionic DNA molecules are condensed into toroidal particulates with the assistance from polycationic PEI molecules (Dunlap et al. 1997). The dielectric properties of the aqueous environment in which they interact with each other could affect the formation and transfection performance of the complex preparations significantly (Ogris et al. 1998). Since the binding of PEI to the immobilized DNA increased with the time of interaction as demonstrated by SPR experiments (Fig. 3b), it is anticipated that thermodynamically more stable complexes can be formed as the result of increasing numbers of engaged electrostatic ion-pairs between them. The presence of competing ions such as Na+ and Cl− is considered as the key to enable spatial re-arrangement between PEI and DNA during complex formation. So it is logical to anticipate the growth of complexes during preparation is simultaneously accompanied by an increasing degree of association between PEI and DNA molecules.
Under the conditions chosen, molecules of PEI and DNA formed complexes in 150 mM NaCl and the complex population experienced a dynamic growth (Fig. 1). To obtain an estimate of the effectiveness of these complexes in gene transfer, FACS analyses of the transfected cells and naked nuclei hypotonically isolated from them were executed immediately after the transfection of 4 h with PEI/Cy5-pEGFP-N1 complexes. With complexes of 321, 449 and 1,734 nm in average diameter, the nuclear presence of pEGFP-N1 in the total population accounted for 76, 70 and 36%, respectively, corresponding to the cellular presence of 90, 75 and 38%. So the complexes clearly benefited from their increased stability to survive the endosomal or lysosomal challenges and subsequently transfer the transgene into the nuclei. In the demonstration of the criticalness of pEGFP-N1 in the complexed form to its nuclear entry, transfection of isolated naked nuclei with PEI/Cy5-pEGFP-N1 complexes yielded a Cy5-pEGFP-N1 positive rate of 79%, in contrast to negligible with free Cy5-pEGFP-N1 under the same conditions. Consequently, an efficient nuclear entry of the transgene after complex endocytosis requires a sufficient degree of association between PEI and DNA molecules for them to remain complexed after their escape from the endosomes/lysosomes. Since the integrity of the complexes is subject to constant challenges from proteins, RNA molecules or chromosomal DNA in the cytoplasmic and/or nuclear environment (Bertschinger et al. 2006), they eventually will transform from the complexed form into the dispersed form as the consequence of their disassembly. As observed from CLSM studies, successful transgene expression seemed only related to cells with co-existence of PEI and DNA in dispersed form in the nuclei. So the disassembly of PEI/DNA complexes to transform them into dispersed forms is critical to the transcription of the transgene. For example, the failure of transforming from PEI/DNA aggregates to a dispersed form in the nuclear region after 24 h of post-transfection culture is believed to be responsible for disabling or delaying GFP expression within the daughter cell on the right (Fig. 5b). Complexes with a lower degree of association may prematurely dissociate in the cytoplasm, consequently disabling the nuclear entry and expression of the transgene. This may exactly explain the observation made to complex preparation of 5 min which exhibited the highest uptake of PEI/pEGFP-N1 but a lower percentage of GFP expressing cells (Fig. 2). The cells with dispersed DNA without the co-existence of dispersed PEI and the appearance of GFP may also fall into this category, i.e., PEI/DNA complexes dissembled intracellularly prior to their nuclear entry because of their low degrees of association (Fig. 6a, the left and right bottom cells; Fig. 6b, the upper left cell).
One of the direct consequences of increased complex stability is the lowered efficiency of endocytosis/phagocytosis (Fig. 2). This is supported by our CLSM observation that the uptake of complexes was mostly restricted to those smaller than 1 μm in size (Fig. 5). Secondly, a greater degree of association may lead to an increased difficulty in complex dissociation and thus a delay of gene transcription. The in vitro transcription assay also clearly told us that the transcription activity was abruptly switched off at a N/P ratio of 1.5–3.1, lower than that required for the completion of complex formation (N/P ratio 3.8–7.7) (Fig. 7). Full or partial dissociation of PEI/DNA complexes is therefore necessary to make the transgene accessible to the nuclear transcription machinery. In agreement with our SPR results (Fig. 3d), complex formation in the absence of NaCl allowed PEI and DNA molecules to compact too tightly to initiate transcription (Ogris et al. 1998). As the time of complex formation increased from 10 to 120 min, the transfection efficiency decreased from 40 to 13% (Fig. 2c). Since the average intracellular levels of pEGFP-N1 in the pEGFP-N1 positive cells were relatively equal among all groups (Fig. 2b), derived data suggested that the apparent activity of protein synthesis in cells (Fig. 2d) and the apparent total activity of both gene transcription and protein synthesis (Fig. 2e) were negatively affected by complex formation time. These differences in the outcome of transfection may suggest that qualities other than the sizes of complex particulates, such as the degree of association, were possibly also playing paramount roles in gene expression. Because intracellular complexes became mostly dispersed within 48 h after transfection (Fig. 6), it is anticipated that endocytosed complexes of a high stability will very likely cause a delay in gene expression. How the rate of complex disassembly affects the overall efficiency of protein expression remains to be determined.
Although 293-F cells discriminated the complex preparations against their sizes during the event of endocytosis/phagocytosis, the average content of pEGFP-N1 in the cells successfully transfected was found to be relatively even among all the groups (Fig. 2b). We may therefore speculate that the cells had a physiologically restrictive tolerance to the intake of PEI/DNA complexes and/or free PEI.
The presence of GFP was detectable by FACS in 1.8% of the total population after a transfection of 4 h (Fig. 4c). This not only tells us the approximate minimal time scale required for the all the events from endocytosis/phagocytosis to mRNA translation, but also the possible existence of heterogeneity in the populations of cells and complexes. Non-viral gene delivery is generally believed to be cell cycle dependent, with the temporary breakdown of nuclear membrane in G2/M phase as the major route for the nuclear entry of plasmids (Tseng et al. 1999; Brunner et al. 2000; Grosjean et al. 2002; Mannisto et al. 2005). The nuclear entry of PEI-complexed DNA was further suggested to be an event of passive importation during mitosis (Grosse et al. 2006). However, results in Fig. 4b clearly indicated the endocytosis of PEI/pEGFP-N1 complexes by 293-F cells was cell cycle independent. The similar distributions of GFP positive 293-F cells and the total population also suggested the possibility of the existence of cell cycle independent mechanism for pEGFP-N1 expression (Fig. 4a, c). Further analyses of the top 20 and 50% of the 293-F cells in GFP intensity also supported this conclusion (data not shown). A similar conclusion was also drawn by transfecting Hela cells in different stages of cell cycle with a linear PEI complexed DNA (Brunner et al. 2002).
PEI-mediated transient gene expression in mammalian cells is an attractive and promising technology for protein production in the biopharmaceutical industry. This study demonstrated the heterogeneous nature of the PEI/DNA complexes by the current method of preparation in terms of the size of the complex particulates and the degree of association between PEI and DNA molecules. These interrelated qualities may have profound effects on the performance of the PEI-mediated transient gene expression processes. Our results may provide valuable information for consideration in the fine-tuning and rational design of PEI-mediated transfection systems in the future.
Acknowledgments
This project was funded by the National High Technology Program (National 863 Plan), Project No. 2007AA021702, the Ministry of Science & Technology, China. The authors wish to thank Mr. Zhao-feng Luo, the University of Science & Technology of China, Hefei, for his outstanding assistance in instrumental analyses.
Footnotes
The authors Xiangzong Han and Qiangyi Fang contributed equally to this work.
References
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA et al (2002) Current protocols in molecular biology. John Wiley & Sons, New Jersey
- Backliwal G, Hildinger M, Chenuet S, Wulhfard S, De Jesus M, Wurm FM (2008) Rational vector design and multi-pathway modulation of HEK 293E cells yield recombinant antibody titers exceeding 1 g/l by transient transfection under serum-free conditions. Nucleic Acids Res 36:e96 [DOI] [PMC free article] [PubMed]
- Baldi L, Muller N, Picasso S, Jacquet R, Girard P, Thanh HP, Derow E, Wurm FM (2005) Transient gene expression in suspension HEK-293 cells: application to large-scale protein production. Biotechnol Prog 21:148–153 [DOI] [PubMed]
- Baldi L, Hacker DL, Adam M, Wurm FM (2007) Recombinant protein production by large-scale transient gene expression in mammalian cells: state of the art and future perspectives. Biotechnol Lett 29:677–684 [DOI] [PubMed]
- Bertschinger M, Backliwal G, Schertenleib A, Jordan M, Hacker DL, Wurm FM (2006) Disassembly of polyethylenimine-DNA particles in vitro: implications for polyethylenimine-mediated DNA delivery. J Control Release 116:96–104 [DOI] [PubMed]
- Boussif O, Lezoualch F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP (1995) A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci USA 92:7297–7301 [DOI] [PMC free article] [PubMed]
- Brunner S, Sauer T, Carotta S, Cotten M, Saltik M, Wagner E (2000) Cell cycle dependence of gene transfer by lipoplex polyplex and recombinant adenovirus. Gene Ther 7:401–407 [DOI] [PubMed]
- Brunner S, Furtbauer E, Sauer T, Kursa M, Wagner E (2002) Overcoming the nuclear barrier: cell cycle independent nonviral gene transfer with linear polyethylenimine or electroporation. Mol Ther 5:80–86 [DOI] [PubMed]
- Capecchi MR (1980) High efficiency transformation by direct microinjection of DNA into cultured mammalian cells. Cell 22:479–488 [DOI] [PubMed]
- Chu YW, Wang R, Schmid I, Sakamoto KM (1999) Analysis with flow cytometry of green fluorescent protein expression in leukemic cells. Cytometry 36:333–339 [DOI] [PubMed]
- Derouazi M, Girard P, Van Tilborgh F, Iglesias K, Muller N, Bertschinger M, Wurm FM (2004) Serum-free large-scale transient transfection of CHO cells. Biotechnol Bioeng 87:537–545 [DOI] [PubMed]
- Dunlap DD, Maggi A, Soria MR, Monaco L (1997) Nanoscopic structure of DNA condensed for gene delivery. Nucleic Acids Res 25:3095–3101 [DOI] [PMC free article] [PubMed]
- Forbes DJ (1992) Structure and function of the nuclear-pore complex. Annu Rev Cell Biol 8:495–527 [DOI] [PubMed]
- Girard P, Porte L, Berta T, Jordan M, Wurm FM (2001) Calcium phosphate transfection optimization for serum-free suspension culture. Cytotechnology 35:175–180 [DOI] [PMC free article] [PubMed]
- Girard P, Derouazi M, Baumgartner G, Bourgeois M, Jordan M, Jacko B, Wurm FM (2002) 100-liter transient transfection. Cytotechnology 38:15–21 [DOI] [PMC free article] [PubMed]
- Godbey WT, Wu KK, Mikos AG (1999) Tracking the intracellular path of poly(ethylenimine)/DNA complexes for gene delivery. Proc Natl Acad Sci USA 96:5177–5181 [DOI] [PMC free article] [PubMed]
- Grosjean F, Batard P, Jordan M, Wurm FM (2002) S-phase synchronized CHO cells show elevated transfection efficiency and expression using CaPi. Cytotechnology 38:57–62 [DOI] [PMC free article] [PubMed]
- Grosse S, Thevenot G, Monsigny M, Fajac I (2006) Which mechanism for nuclear import of plasmid DNA complexed with polyethylenimine derivatives? J Gene Med 8:845–851 [DOI] [PubMed]
- Han X, Sun L, Fang Q, Li D, Gong X, Wu Y, Yang S, Shen BQ (2007) Transient expression of osteopontin in HEK 293 cells in serum-free culture. Enzyme Microb Technol 41:133–140 [DOI]
- Kopatz I, Remy JS, Behr JP (2004) A model for non-viral gene delivery: through syndecan adhesion molecules and powered by actin. J Gene Med 6:769–776 [DOI] [PubMed]
- Kunath K, von Harpe A, Fischer D, Peterson H, Bickel U, Voigt K, Kissel T (2003) Low-molecular-weight polyethylenimine as a non-viral vector for DNA delivery: comparison of physicochemical properties, transfection efficiency and in vivo distribution with high-molecular-weight polyethylenimine. J Control Release 89:113–125 [DOI] [PubMed]
- Löfås S, Johnsson B (1990) A novel hydrogel matrix on gold surfaces in surface plasmon resonance sensors for fast and efficient covalent immobilization of ligands. J Chem Commun 21:1526–1528
- Lungwitz U, Breunig M, Blunk T, Gopferich A (2005) Polyethylenimine-based non-viral gene delivery systems. Eur J Pharm Biopharm 60:247–266 [DOI] [PubMed]
- Mannisto M, Ronkko S, Matto M, Honkakoski P, Hyttinen M, Pelkonen J, Urtti A (2005) The role of cell cycle on polyplex-mediated gene transfer into a retinal pigment epithelial cell line. J Gene Med 7:466–476 [DOI] [PubMed]
- Ogris M, Steinlein P, Kursa M, Mechtler K, Kircheis R, Wagner E (1998) The size of DNA/transferrin-PEI complexes is an important factor for gene expression in cultured cells. Gene Ther 5:1425–1433 [DOI] [PubMed]
- Schlaeger EJ, Christensen K (1999) Transient gene expression in mammalian cells grown in serum-free suspension culture. Cytotechnology 30:71–83 [DOI] [PMC free article] [PubMed]
- Sun X, Goh PE, Wong KT, Mori T, Yap MG (2006) Enhancement of transient gene expression by fed-batch culture of HEK 293 EBNA1 cells in suspension. Biotechnol Lett 28:843–848 [DOI] [PubMed]
- Sun X, Hia HC, Goh PE, Yap MG (2008) High density transient gene expression in suspension-adapted 293 EBNA1 cells. Biotechnol Bioeng 99:108–116 [DOI] [PubMed]
- Thomas M, Klibanov AM (2003) Non-viral gene therapy: polycation-mediated DNA delivery. Appl Microbiol Biotechnol 62:27–34 [DOI] [PubMed]
- Tseng WC, Haselton FR, Giorgio TD (1999) Mitosis enhances transgene expression of plasmid delivered by cationic liposomes. Biochim Biophys Acta 1445:53–64 [DOI] [PubMed]
- Ungaro F, De Rosa G, Miro A, Quaglia F (2003) Spectrophotometric determination of polyethylenimine in the presence of an oligonucleotide for the characterization of controlled release formulations. J Pharm Biomed Anal 31:143–149 [DOI] [PubMed]
- Werth S, Urban-Klein B, Dai L, Hobel S, Grzelinski M, Bakowsky U, Czubayko F, Aigner A (2006) A low molecular weight fraction of polyethylenimine (PEI) displays increased transfection efficiency of DNA and siRNA in fresh or lyophilized complexes. J Control Release 112:257–270 [DOI] [PubMed]
- Wurm FM (2004) Production of recombinant protein therapeutics in cultivated mammalian cells. Nat Biotechnol 22:1393–1398 [DOI] [PubMed]