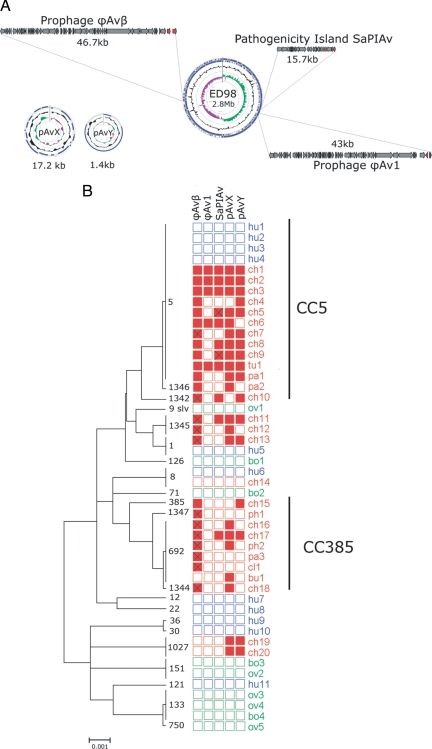
Fig. 2.
S. aureus avian isolates of distinct clonal lineage share a common accessory gene pool. (A) Novel MGEs identified in the whole-genome sequence of the ST5 poultry isolate ED98. The ED98 genome and plasmid maps represent ORFs (blue arrows on outer circle), GC content (middle circle), and GC skew (inner circle; GC+ and GC- skew in green and purple, respectively). Prophage and SaPI ORFs are represented by arrows, and red arrows denote genes putatively involved in virulence and/or host specificity. (B) The distribution of novel MGEs identified in the ED98 genome sequence among a panel of diverse isolates of avian, human, bovine, and ovine origin. Neighbor joining tree with bootstrapping consensus inferred from 500 replicates was constructed using concatenated sequences of S. aureus STs from different host species, including representatives of the major clonal complexes identified in the S. aureus species. Labels at branch tips denote STs of strains. A filled box indicates presence of an MGE and an empty box its absence as determined by MGE-specific PCR. A crossed box indicates the presence of a related but non-identical genetic element (determined by successful amplification in 1 of 2 PCRs). Red, blue, and green boxes denote strains of avian, human, and ruminant (bovine or ovine) origin, respectively. Strain prefixes denote the host species origin of the strains including ch, broiler chicken; ph, farmed pheasant; tu, farmed turkey; pa, farmed partridge; cl, poultry layer; bu, wild buzzard; hu, human; bo, cattle; and ov, sheep, respectively.