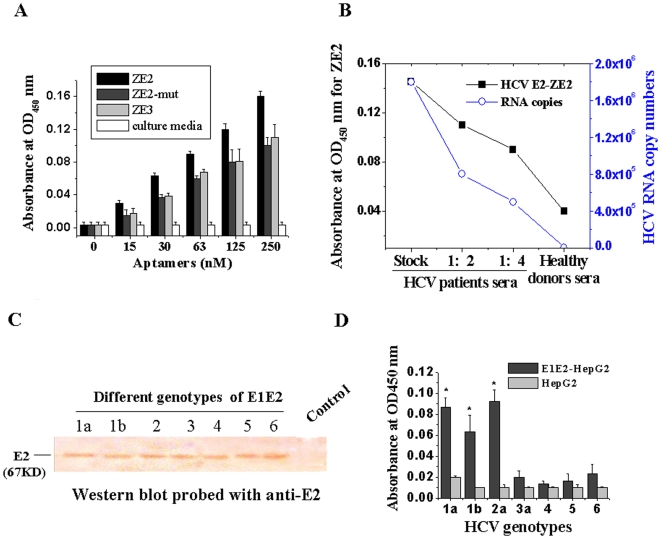
Figure 4. The ssDNA aptamer ZE2 specifically targets HCV particles.
(A) HCVcc could be captured by the aptamer ZE2 in an aptamer dose-dependent manner by sandwich ELISA. Plates were coated with anti-E2 polyclonal antibody and blocked with 1% BSA. After extensive washing, the bound viral particles were added and then revealed using biotin-labeled aptamer ZE2, ZE3 or ZE2-mut as described in Materials and Methods. Data shown were calculated as mean±SEM, and data are from three independent experiments. (B) Comparison of HCV detection methods: HCV E2 antigen-aptamer method and HCV RNA quantification by real time fluorescence quantitative RT-PCR. The aptamer method was performed as above (A), except HCV patients and healthy donor serum samples were added into each 96-well ELISA plate instead of HCVcc. (C) Determination of E2 protein expressions of different genotypes of E1E2 gene stable-expressing HepG2 cells with anti-E2 antibody by western blot analysis. HepG2 was uesd as a control. (D) Different genotypes of E2 detected by aptamer ZE2. Data shown were calculated as mean±SEM and data are from three independent experiments.