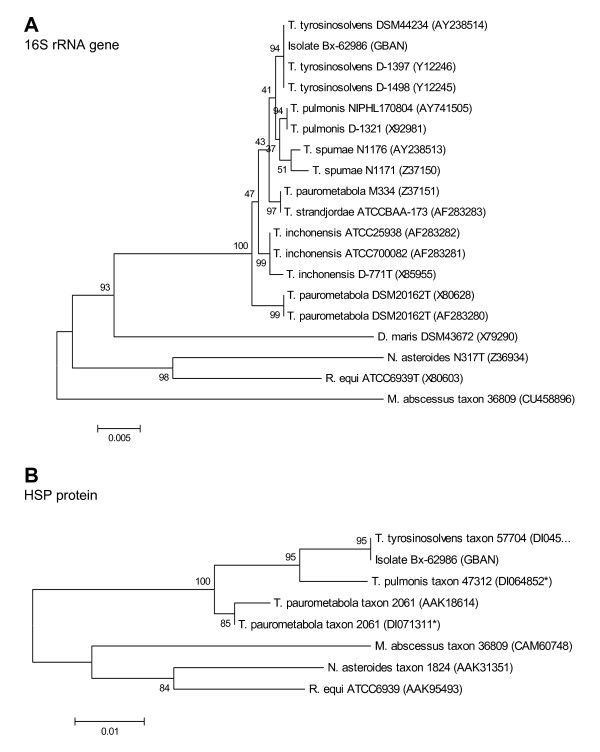
Figure 1.
Phylogenetic analysis of 16S rRNA and HSP generated with the neighbor-joining method. The phylogeny presented is based on the alignment of 1341 bp of the 16S rRNA and 201 residues of the HSP sequences. Theses consensus trees were rooted on the sequences of Rhodococcus equi, Nocardia asteroides, Mycobacterium abscessus and Dietzia maris (except for the HSP sequence not available for this species). Numbers next to each node indicate bootstrap values as a percentage of 1000 replicates. The bootstrap values below 50% are indicated by a slash (/). Each strain designation is followed by the corresponding Genbank accession number in parentheses. Asterisks indicate a Genbank accession numbers corresponding to a nucleotide sequence whose amino acid sequence was deduced.