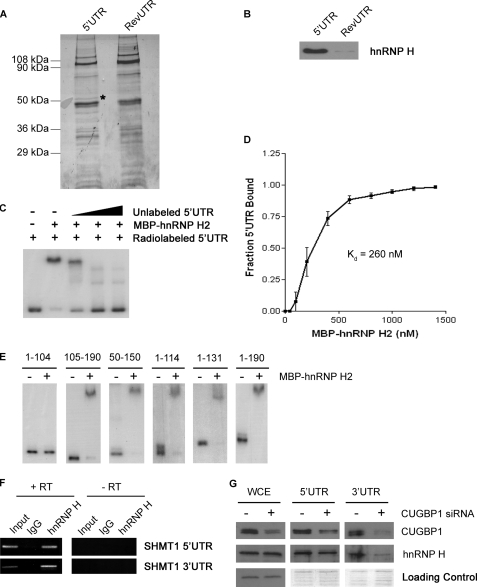
FIGURE 4.
Interaction of hnRNP H2 with the SHMT1 UTRs. A, MCF-7 whole cell extract was applied to RNA affinity columns to which either the SHMT1 5′-UTR or the RevUTR had been attached. Proteins that bound to the UTRs were eluted, separated by SDS-PAGE, and visualized by Coomassie Blue staining. The protein marked with an asterisk was excised from the gel and analyzed by μLC/MS/MS. B, the SDS-polyacrylamide gel from A was transferred to a PVDF membrane and analyzed by Western blotting using an antibody against hnRNP H. C, electrophoretic mobility shift assays were carried out in the presence of excess yeast tRNA to eliminate nonspecific binding of the probe, radiolabeled SHMT1 5′-UTR, and recombinant MBP-hnRNP H2. A 10×, 50×, and 100× molar excess of unlabeled SHMT1 5′-UTR was also added in lanes 3, 4, and 5, respectively, to determine binding specificity. D, electrophoretic mobility shift assays were carried out using radiolabeled SHMT1 5′-UTR and increasing concentrations of MBP-hnRNP H2. The fraction of the 5′-UTR bound by the recombinant protein was quantified using ChemiImager 4400 from Alpha Innotech Corp. The dissociation constant (Kd) was determined using GraphPad Prism. E, electrophoretic mobility shift assays were carried out using radiolabeled SHMT1 5′-UTR truncation mutants in the absence and presence of MBP-hnRNP H2. The nucleotides of the 5′-UTR that comprise the truncation mutant are listed above each gel. The nucleotide at the 5′-end of the 5′-UTR is labeled 1. The nucleotide at the 3′-end of the 5′-UTR is labeled 190. F, RNA was immunoprecipitated from UV-treated MCF-7 whole cell extract using an antibody against IgG (control for nonspecific binding) or hnRNP H. The RNA in lanes 1–3 (+RT) was reverse-transcribed into cDNA and then analyzed by PCR using primers specific to either the SHMT1 5′-UTR or the SHMT1 3′-UTR. The RNA in lanes 4–6 (−RT) did not undergo the reverse transcription step. Rather, they were analyzed directly by PCR to control for DNA contamination in the immunoprecipitates. Lanes 1 and 4 (Input) represent 1% of the RNA used in the immunoprecipitation. G, whole cell extract (WCE) from cells that were treated with either negative control siRNA (−CUGBP1 siRNA) or siRNA directed against CUGBP1 were applied to RNA affinity columns to which either the cSHMT 5′-UTR or the cSHMT 3′-UTR had been attached. Proteins that bound to the UTRs were eluted, separated by SDS-PAGE, and visualized by Western blotting using antibodies against CUGBP1 or hnRNP H. For the whole cell extract, equal protein loading was confirmed by using an antibody against GAPDH. For the affinity column elutions, equal protein loading was confirmed by staining the gel with Coomassie blue after transfer.