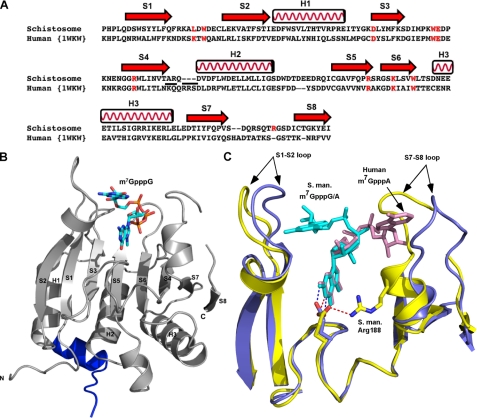
FIGURE 2.
Comparison of schistosome eIF4E with human eIF4E shows overall similarity in structure but differences in the orientation of the second base in the cap. A, structure-based sequence alignment of schistosome eIF4E with human eIF4E. The secondary structural elements were assigned based on the schistosome eIF4E structure. Residues involved in cap binding are colored in red. The underlined residues are substitutions and deletions discussed in the text. The alignment was prepared using the combinatorial extension algorithm (68) and an on-line server. B, the overall view of schistosome eIF4E structure. The 4E-BP peptide is colored blue. Three α-helices and eight β-strands are labeled. The m7GpppG cap is shown as sticks. C, superimposition of schistosome and human eIF4E complexes bound to the dinucleotide cap. Note the difference in orientation of the second cap base (marked by arrows) between schistosome eIF4E (yellow) and human eIF4E complex structure (Protein Data Bank code 1WKW) (blue). The S1-S2 and S7-S8 loops and caps are marked. The position of the key residue Glu-90 (schistosome, yellow; human, blue color) is also shown. The colored dashes (schistosome, red; human, blue) denote a hydrogen bond between the N2 amide from the N-7 methylated guanine base and the side chain of Glu. In addition, one hydrogen bond is shown between the side chain of Glu-90 to Nη of Arg-188 in the schistosome eIF4E complex structure. All the pictures were generated using Pymol (44). S. man., S. mansoni.