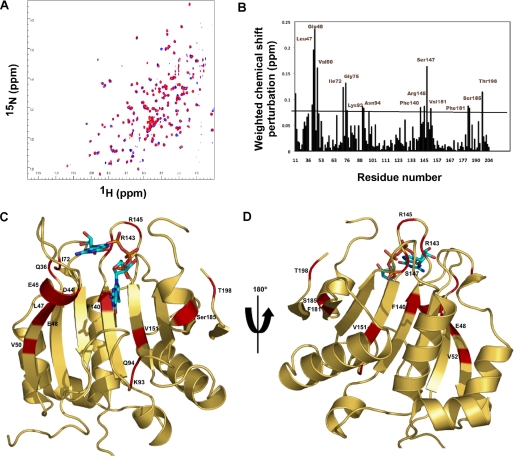
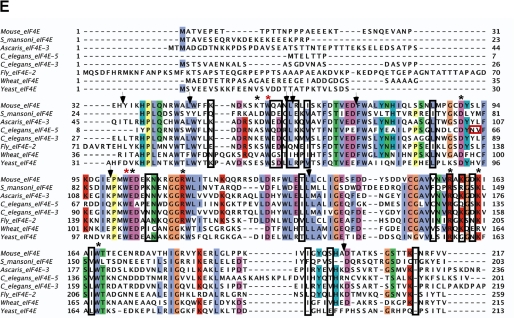
FIGURE 5.
NMR-determined chemical shift perturbations on schistosome eIF4E binding to m2,2,7GpppG compared with m7GpppG. A, superposition of the 1H-15N HSQC spectrum of the schistosome eIF4E bound to m7GpppG (red) and bound to m2,2,7GpppG (blue). B, specific chemical shift perturbations on binding m2,2,7GpppG compared with m7GpppG. The reported chemical shift Δδ represents the weighted chemical shift by applying the Pythagorean theorem: Δδ{1H,15N} = {Δδ(1H)2 + 0.2 × Δδ(15N)2}½ where Δδ(1H) and Δδ(15N) are the chemical shift difference of the amide proton and nitrogen, respectively (28, 69). Amino acid residues with large chemical shifts are labeled. C and D, residues with major chemical shift perturbation on binding m2,2,7G compared with the m7G cap mapped onto the crystal structure of the m7GpppG·eIF4E complex. The backbone residues with major chemical shifts are colored red. Residues with major chemical shifts in the vicinity of the second base of the cap are shown in C, and those with major chemical shifts in the vicinity of the third phosphate of the cap are shown in D. E, Clustal sequence alignment of several eIF4E illustrating conserved residues, residues undergoing CSP, and mouse pepsin cleavage sites on cap binding. The black boxes denote residues with major chemical shift perturbation in our NMR experiments as described above. The vertical black arrows denote the specific pepsin cleavage sites based on the mammalian eIF4E (21). Starred residues are those involved in cap binding with the two key Trp residues in red. The red box indicates two key residues for C. elegans eIF4E reported to be important for m2,2,7G cap binding specificity (24). Note that C. elegans eIF4E-5 and Ascaris eIF4E-3 binds both types of caps, whereas mouse eIF4E3 and C. elegans eIF4E-3 are specific for m7G cap.