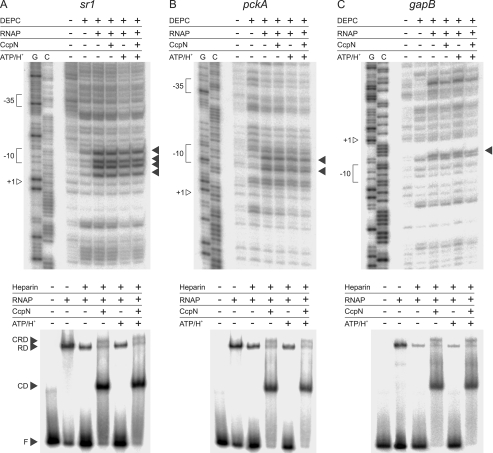
FIGURE 2.
Open complex formation assay at the sr1 (A), pckA (B), and gapB (C) promoters. Probing with DEPC is shown at the top while the corresponding heparin-probing is shown below. For DEPC-probing, DEPC (10%), RNAP (100 nm), CcpN-His5 (100 nm), were added where indicated. Bands showing the presence of single-stranded DNA regions, and therewith open complexes are indicated by arrows. G, G→A sequencing reaction, C, C+T sequencing reaction. Positions of +1, the −10 and −35 box are indicated. Please note that the noncoding strand was used for sr1 and pckA, while the coding strand was used for gapB. For heparin-probing, Heparin (0.1 g/liter) CcpN-His5 (3 μm), RNAP-His6 (3 μm), ATP (3 mm), or HCl (to a final pH of 6.5) were added where indicated. F, free DNA; CD, CcpN-DNA complex; RD, RNAP-DNA complex; CRD, CcpN-RNAP-DNA complex. Autoradiograms of the gels are shown.