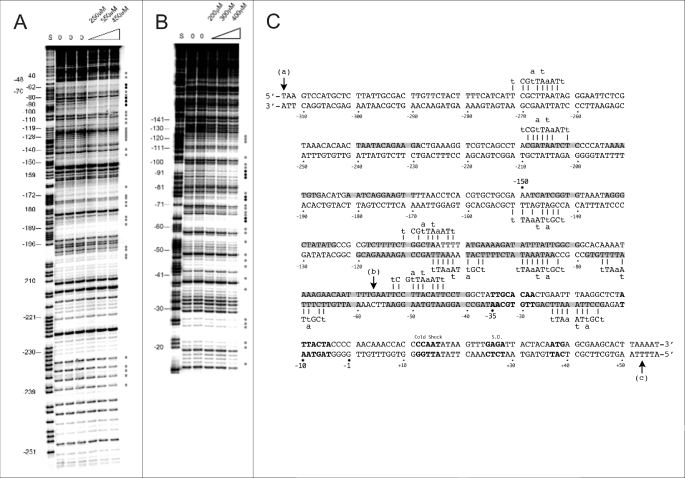
FIGURE 3.
Protection of the hns promoter by H-NSctd. A and B, electrophoretic separation of the fragments obtained following DNaseI footprinting (see “Experimental Procedures”) of the coding (A) and noncoding (B) strands of the hns promoter. Protected nucleotides are indicated by gray or black dots depending upon the intensity of the protection. C, sequence of the hns promoter fragment cloned into pKK400. The numbers indicate nucleotide positions with respect to the transcription start. The sites affected by H-NSctd are highlighted in gray, whereas the −35 and −10 core elements of the promoter, the cold shock recognition signal, the Shine-Dalgarno sequence (S.D.), and the initiation triplet are indicated (from top left) in bold. The H-NS-binding motifs are indicated above or below the DNA duplex, and the short vertical bars indicate the base match between motifs and promoter sequence. Vertical arrows (a) and (b) indicate the 5′ ends of the 400-bp DNA promoter fragment present in pKK400 and of the 110-bp fragment in pKK110, respectively, whereas arrow (c) indicates the 3′ terminus of both fragments.