Abstract
We have screened an odorant compound library and discovered molecules acting as chemical signals that specifically activate both G-protein-coupled olfactory receptors (ORs) on the cell surface of olfactory sensory neurons and the human nuclear estrogen receptor α (ER) involved in transcriptional regulation of cellular differentiation and proliferation in a wide variety of tissues. Hence, these apparent dual active odorants induce distinct signal transduction pathways at different subcellular localizations, which affect both neuronal signaling, resulting in odor perception, and the ER-dependent transcriptional control of specific genes. We demonstrate these effects using fluorescence-based in vitro and cellular assays. Among these odorants, we have identified synthetic sandalwood compounds, an important class of molecules used in the fragrance industry. For one estrogenic odorant we have also identified the cognate OR. This prompted us to compare basic molecular recognition principles of odorants on the two structurally and apparent functionally non-related receptors using computational modeling in combination with functional assays. Faced with the increasing evidence that ORs may perform chemosensory functions in a number of tissues outside of the nasal olfactory epithelium, the unraveling of these molecular ligand-receptor interaction principles is of critical importance. In addition the evidence that certain olfactory sensory neurons naturally co-express ORs and ERs may provide a direct functional link between the olfactory and hormonal systems in humans. Our results are therefore useful for defining the structural and functional characteristics of ER-specific odorants and the role of odorant molecules in cellular processes other than olfaction.
Our nose detects a large variety of odorant signals relying on about 350 different predicted G-protein-coupled olfactory receptors (ORs)3 with a conserved seven-transmembrane helical structure (1–3). By binding odorant molecules ORs modulate the conversion of chemical into electrical neuronal signals that are finally decoded in higher brain regions to trigger emotional and behavioral responses (4–6). Odorants are generally small, hydrophobic organic molecules with highly variable chemical structures and properties (7). They easily pass cell membranes, and, due to their enormous chemical diversity, some of them might, apart from their “conventional” role in olfaction, trigger also yet unknown cellular processes. The role of specific ORs in sperm chemotaxis has been documented (8, 9), and the widespread ectopic expression of OR genes in a large number of non-olfactory human tissues implicates additional, unproven functions of ORs in embryonic development and cell-cell recognition (10), and proliferation rates in prostate cancer cells (11). Recent findings indicate that ORs might also have chemosensory functions in kidney (12). Although most present efforts concentrate on matching odorants with their cognate ORs to define their molecular receptive ranges and receptor functions (13–17), the screening for particular functions of odorants as intracellular signals remains in its infancy.
Here we report that certain natural and synthetic odorants (see Fig. 1), commonly used as fragrances in cosmetic articles, activate both ORs at the cell surface and intracellular human estrogen receptor α (ER) predominantly localized in the cell nucleus. The activation of endogenous ER by the odorants was assessed on human MCF7 breast cancer cells representing a highly sensitive and widely accepted assay system for detecting estrogenic activities of natural or synthetic compounds (18–20). ER is a prototypical member of the ligand-inducible transcription factors, known to perform multiple molecular interactions according to external chemical stimuli (21).
FIGURE 1.
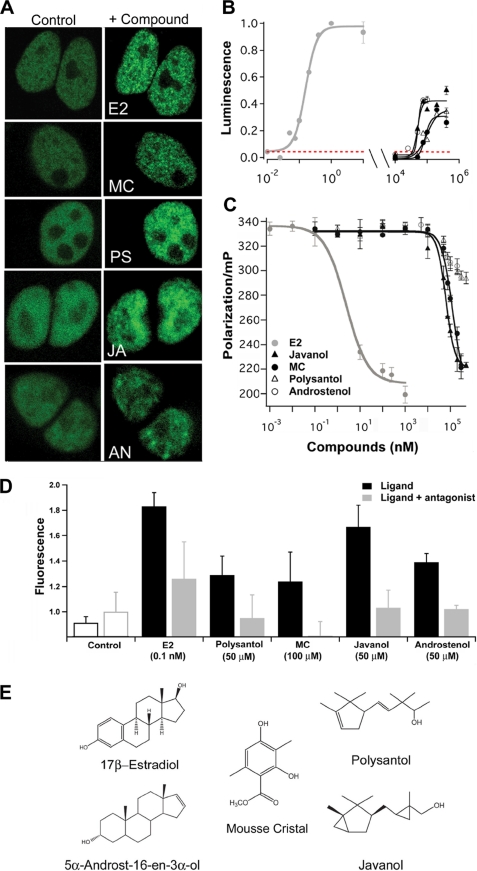
Estrogenic odorant compounds. A, fluorescence confocal micrographs showing odorant-dependent intranuclear clustering of ER-YFP in HEK293 cells (ex. 514 nm, em. 530 nm). Nuclear distribution of ER-YFP before addition of odorant (left panels), and 15 min after addition (right panels), of 10 nm natural cognate ligand E2 (E2 panel), 100 μm MC (MC panel), 100 μm Polysantol (PS panel), 100 μm Javanol (JA panel), and 10 μm Androstenol (AN panel). B, activation of endogenous ER by estradiol and different odorants monitored by luciferase reporter gene expression. MCF7 cells were co-transfected with a p2xERE-Luciferase reporter, and a Renilla luciferase expression vector for normalization of the cellular luminescence responses. Compounds were added 8 h after transfection and incubation at 37 °C was continued for 20 h. Luminescence was measured using GloMax 96 Microplate Luminometer (Promega). The co-application of ER-specific antagonist ICI 182,780 together with increasing concentrations of the relevant odorant compounds completely blocked the ER activation in the reporter assay. Representative results are shown for MC (red dashed line). C, competitive binding of odorants to ER determined by fluorescence polarization. Concentration-dependent displacement of 1 nm fluorescent estrogen ligand (FluormoneTM ES2, dissociation constant Kd = 4 ± 2 nm) from ER (15 nm) by increasing concentrations of the indicated compounds. Points are mean values of triplicate measurements, expressed as millipolarization (mP) units. D, histograms show the proliferation of MCF7 cells 4 days after application of E2 and the odorant compounds at the indicated concentrations (n = 6). The co-application of the ICI 182,780 (10 μm) significantly reduced proliferation induced by E2 and the odorants. Controls were either not treated with ligands (black line) or treated with ICI 182,780 at 10 μm (gray line). E, chemical structures of the natural cognate ligand estradiol and of estrogenic odorants.
Activated by its natural agonist 17β-estradiol (E2), ER triggers transcription of target genes involved in various biological functions like cell growth and differentiation (22). An increasing number of structurally diverse estrogenic compounds have been identified and proposed as risk factors for disruption of reproductive development and tumorigenesis (23). Estrogenic chemicals are defined as substances, interacting and activating ER, which may interfere with the endocrine system (24). Here we report activating effects of odorants on ER due to the ability of ER to bind a wide variety of small hydrophobic compounds. These investigations contribute to the unraveling of general molecular principles and consequences of odorant interactions with different cellular targets.
Several cellular targets for E2 have been discovered in recent years. Obviously this steroid hormone mediates its physiological effects in a combined action of nuclear receptors, ER α and ER β, and a membrane-localized G-protein-coupled receptor GPR30 indicating that small lipophilic molecules may function as chemical signals at various subcellular localizations (25). In analogy, hydrophobic odorant molecules, which are cell-permeable, might also be capable of interacting with different cellular targets.
EXPERIMENTAL PROCEDURES
Materials
Odorants (from Givaudan, Switzerland, and Sigma, Switzerland) were freshly dissolved in DMSO before experiments.
Cell Culture
MCF7 human breast cancer cells (gift from Ana Soto, Tufts University, Boston) were grown in Dulbecco's modified Eagle's medium (DMEM, Invitrogen) supplemented with 5% fetal bovine calf serum (Invitrogen, Carlsbad, CA) at 37 °C in 5% CO2 atmosphere and saturating humidity.
Site-directed Mutagenesis
Mutations were introduced into mOR-EG cDNA using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). All of the point mutation products were digested with HindIII and NotI and inserted into the vector pCEP4 (Invitrogen). All mutations were confirmed by DNA sequencing.
Investigating Activation of OR
The cDNA of the mouse mOR-EG (26) comprising a 5′ DNA extension encoding the N-terminal 20 amino acids of bovine rhodopsin (13) was subcloned as a HindIII/NotI restriction fragment into the mammalian expression vector peak8 (Edge Biosystems) and designated pRhoOR-EG. Cellular responses after addition of odorants were measured in 96-well plates (Greiner, Germany) containing 35,000 Hana3A cells (27) per well co-transfected with 150 ng of pRhoOR-EG and 150 ng of cAMP-response element secreted alkaline phosphatase reporter plasmid using Lipofectamine 2000 (Invitrogen). Cells were incubated for 16 h at 37 °C in cell medium with or without test compounds. An aliquot of supernatant from each well was then mixed with an equal volume of 1 m diethanolamine bicarbonate, pH 9.8, containing 20 mm para-nitropheny1 phosphate and 1 mm MgCl2 (Sigma, Switzerland); absorbance was measured at 410 nm using a multiwell plate reader (SpectraMax 360, Molecular Devices). The immunoblot analysis showed single bands representing c-Myc-tagged mOR-EG wild-type and mutant receptors and revealed no obvious difference in the protein expression level (data not shown).
ER Nuclear Redistribution Assay
HEK293 cells adapted to hormone-free DMEM (Invitrogen) supplemented with 10% of charcoal dextran-treated calf serum (HyClone) were seeded into eight-well chambers (Lab-Tek) and transfected with pERα-YFP (21) using Lipofectamine 2000. 16–22 h after transfection, cells were observed by fluorescence confocal microscopy (LSM 510, Zeiss, Germany). Cells exhibiting a homogeneous nuclear distribution of ER-YFP (excitation: 514 nm, emission: >530 nm) were stimulated with odorant solutions and imaged during 45 min at 5-min intervals.
ER Activation Assay
The Dual-Glo System (Promega, Madison, WI) allowed independent measurements of stable luminescence from two reporter genes: the p2xERE-Luc encoding the firefly luciferase (courtesy of B. Desvèrgne, Center for Integrative Genomics, University of Lausanne, Switzerland) was used to quantify ER activation. Renilla luciferase driven by a constitutively active HSV-TK promoter (pGL4.74[hRLuc/TK], Promega) was used as an internal control to normalize the results. 17β-Estradiol (E2) was from Sigma (Switzerland). MCF7 cells were seeded into white 96-well plates (PerkinElmer Life Sciences) at a density of 30,000 cells/100 μl of DMEM, supplemented with 10% of charcoal dextran-treated calf serum (HyClone), and transfected twice using Lipofectamine 2000 at a 5-h interval between the first and second transfection. Briefly, 0.3 μg of DNA and 0.5 μl of Lipofectamine 2000 were each diluted into 25 μl of Opti-MEM medium (Invitrogen) and 30 s later combined for cell transfection. The DNA was a mixture containing p2xERE-Luc, pGL4.74, at a 1:1 ratio. 8 h after the second transfection, cells were incubated for 20 h with E2 or odorant compounds or co-applications of the ER-specific antagonist fulvestrant (ICI 182,780) together with the same ranges of odorant concentrations as used for dose-dependent activation of ER (Fig. 1). Estrogenic effects were determined by quantifying firefly luciferase and Renilla luciferase following the manufacturer's protocol. Luminescence was measured using GloMax 96 Microplate Luminometer (Promega).
ER Ligand Binding Assay
A fluorescence polarization-based assay was used to measure competitive ligand binding to human estrogen receptor α (ERα Competitor Assay, Invitrogen). Serially diluted odorant compounds were added to compete with a fluorescent estrogen ligand (FluormoneTM, ES2, Kd = 4 ± 2 nm) for binding to ER. In 96-microwell plates (Fluotrac 200, Greiner, Germany), 50 μl of ER·ES2 complex was prepared such that the final concentration of ER was 15 nm and the concentration of ES2 was 1 nm and added to 50 μl of screening buffer containing the test compounds at the appropriate concentrations. Competition for ER binding was allowed to come to equilibrium for 2 h at room temperature. Fluorescence polarization was measured using an AnalystTM Assay Detection System (LJL Biosystems) with excitation at 485 nm and emission at 530 nm. IC50 values were determined after fitting the experimental data with the Hill equation using Igor Pro Software (WaveMetrics).
FCS
To analyze the effect of odorant molecules on the mobility of the estrogen receptor, fluorescence correlation spectroscopy (FCS) measurements were performed in HEK cells transfected with ER-YFP. FCS analyses the mobility of fluorescent ER at nanomolar concentrations, which is close to native expression levels of ER. 30 min after adding a potential activating ligand, 100-s fluorescence traces were recorded in the nuclei of transfected cells. A commercial FCS setup (Confocor 3, Carl Zeiss, Jena, Germany) was used. Only the last 80-s periods were used to evaluate auto-correlation curves, to avoid artifacts from photo-bleaching of immobile particles. The ER is involved in a complex interaction network, resulting in a variety of mobile species (21). A detailed investigation of changes caused by odorants in this network will be the scope of future work. Here we only analyze the average mobility of the labeled estrogen receptors, which is greatly reduced upon incubation with ER specific ligands and odorants (supplemental Fig. S1).
Cell Proliferation
MCF7 cells were plated in 96-well plates at initial concentrations of 5 × 103 cells/well in 5% charcoal-treated fetal bovine serum in DMEM). After 24 h at 37 °C with 5% CO2 the seeding medium was removed and replaced by the experimental medium (phenol red-free DMEM 5% charcoal dextran-treated human serum from Sigma) and a range of concentrations of the relevant odorants or mixtures of odorants and the ER-specific antagonist ICI 182,780 at 10 μm were added to the medium. The assay was terminated on day 4 by removing the cell media from the wells and freezing the plates at −80 °C. Cell proliferation was investigated using the CyQUANT cell proliferation kit (Invitrogen).
Quantitative Structure Activity Relationships (QSAR) Analysis
The three-dimensional structures of odorant molecules were generated and energy-minimized in water using the AMBER force field (28) implemented in Macro-Model (29). Atomic partial charges and solvation energies were calculated using the program AMSOL (30). Ligand molecules were flexibly docked to the ER ligand binding site of the crystal structure (31) using the software Yeti (32). Docking poses were visually inspected, and up to four different energetically favorable conformations per molecule were selected as input for the Quasar program. IC50 values for the different ligands were estimated based on the 6D-QSAR model of ER as implemented in Quasar software (33). The resulting IC50 values (E2, 5.35 ± 3.33 nm; Mousse Cristal (MC), 54.7 ± 36.69 μm; Polysantol (R,S), 67.49 ± 11.90 μm; Polysantol (S,S), 772 ± 486 μm; Polysantol (S,R), 216 ± 122 μm; Polysantol (R,R), 2.86 ± 4.45 mm; Javanol (R,R), 11.96 ± 10.55 μm; Javanol (R,S), 2.41 ± 2.64 mm; Javanol (S,S), 176.7 ± 567 μm; Javanol (S,R), 40.73 ± 23.33 μm; and 5α-androst-16-en-3-ol, 25.05 ± 12.5 μm) were used to calculate Ki binding affinities based on the Cheng-Prusoff equation (34). The Kd value for the estrogen receptor was taken from Reese et al. (35), and the ligand concentration was set to 1 × 10−9 m as reported before (36) for the data set used to compute the 6D-QSAR model of ER (33).
OR Ligand-receptor Model
Homology models for the mouse eugenol receptor were built based on the high resolution crystal structures of β2-adrenergic receptor (38). A structure-guided alignment based on β2-adrenergic receptor and bovine rhodopsin (39) was used to generate a multiple sequence alignment between the mouse eugenol receptor (UniProt AC: Q920P2) and several orthologues using 3D_COFFEE (40). This initial target-template alignment was further modified to incorporate a Bayesian estimate of pairwise residue interactions based on correlation between positions of the multiple sequence alignment (41), GPCR family-specific fingerprints (42), functional mutation studies by Katada et al. (26), and the effects of site-directed point mutations. Model coordinates were generated using MODELLER (43) based on several alternative target-template alignments.
MC and eugenol were flexibly docked into each of the alternative receptor models with restraints on hydrogen-bonding interactions on Ser-113 using the Glide Induce Fit Protocol (44) of the Schrödinger Suite 2007. Prior to docking the receptor was checked for correct orientation of Asn and Gln side chains, and both receptor and ligand were assigned ionization and tautomerization states. The receptor was subsequently subjected to restrained optimization based on the OPLS force field (version 2005). The Induced Fit Protocol includes: (i) an initial step of docking using a softened potential (van der Waals radii scaling), (ii) a round of binding site residues side-chain prediction for each of the protein·ligand complexes, (iii) protein minimization of the same set of residues and the ligand for each protein·ligand complex pose, (iv) glide redocking of each protein·ligand complex structure within a specified energy of the lowest energy structure (30 kcal/mol), and finally (iv) estimation of the binding energy (IFDScore) for each result pose.
RESULTS
We screened a large library of odorants with diverse chemical structures for their potential to activate ER. For achieving efficient throughput for detecting estrogenic activities we used fluorescence microscopy to first analyze changes in the intranuclear distribution of a functional ER-YFP chimera, obtained by fusing ER to the yellow fluorescent protein YFP, and then to investigate receptor mobility. Previous studies have shown that ER-activating compounds alter the nucleoplasmic distribution of fluorescent ER chimera from a diffuse to a punctuate pattern (14, 45–48), which correlates with mobility changes of ER (measured by fluorescence recovery after photobleaching and FCS) due to interactions with nuclear DNA and multiple proteins (21, 49–53). We discovered two structurally different types of odorants, which induce within minutes after application nuclear aggregation of ER (Fig. 1A) and substantial reduction of ER mobility, measured by FCS (see supplemental Fig. S1), similar effects as reported for the natural agonist estradiol (54). Most potent for activating ER were methyl 2,4-dihydroxy-3,6-dimethylbenzoate (also known as Mousse Cristal (MC)) and molecules of the sandalwood odorant family, which are extensively used in modern perfumery as synthetic substitutes of the sandalwood oil (54) (Fig. 1E).
A reporter assay based on the endogenously expressed ER in MCF7 breast cancer cells and a transfected firefly luciferase under the transcriptional control of two estrogen-responsive elements was used to distinguish between antagonistic and agonistic properties. Stimulation of ER with E2 was the reference to evaluate the efficacy of the odorants on ER-mediated transcription. MC, the sandalwood odorants Javanol and Polysantol, and the porcine pheromone androstenol (55), exhibiting a musk- and sandalwood-like odor, elicited a robust full dose response in the range of 10−5 to 10−4 m (Fig. 1B), whereas the co-administration of ICI 182,780 reverses the agonistic effect of the odorants. This is the first demonstration that sandalwood-derived odorants act as ER-specific agonists. Estrogenic activity has so far only been reported for a distinct class of odorants: the polycyclic musks (56). We also confirmed the estrogenic activity of the odorant molecules by measuring their proliferative effect on the division of MCF7 human breast cancer cells (19). Odorants at concentrations close to their EC50 values from the reporter assay induced a significant increase in cell yield, which was reduced by co-application of the ER specific antagonist ICI 182,780 (Fig. 1D).
Furthermore, the specificity of the interaction with ER could be confirmed by competitively replacing a fluorescent estrogenic ligand in a dose-dependent manner by different odorants as determined by fluorescence polarization measurements (Fig. 1C). The fluorescent estrogen-ligand bound to ER exhibits a high fluorescence polarization value, whereas displacement of the fluorescent estrogen ligand from ER by the corresponding odorant decreases the fluorescence polarization in a dose-dependent manner, which is a well established assay to probe the specific binding of molecules to ER (57, 58). Although Polysantol and androstenol specifically competed for the ER binding site, we were not able to reach the same baseline level of ligand displacement as for the other compounds (Fig. 1C) due to solubility problems of these odorants at concentrations >100 μm. The Ki values are very close to the EC50 values of the transactivation responses (Table 1) with the exception of E2, for an unknown reason. Such differences have also been documented for other estrogenic compounds that induce transactivation in the micromolar range (59). Overall our experimental binding data are in good agreement with the in silico odorant affinity calculations (Table 1). For Javanol and Polysantol docking and binding affinity estimations were performed separately for each stereoisomer of a particular odorant, whereas experiments were performed using mixtures of different stereoisomers. Our findings indicate that exogenously added odorants are capable of entering the cell nucleus and, depending on their chemical structure, may act as agonists of ER.
TABLE 1.
Estrogenic activities of odorants
The compound concentrations EC50 evoking half-maximal estrogenic activity in transcription activation, the dissociation inhibition constants Ki calculated from the IC50 values of the ES2 displacement curves (Fig. 1), and the corresponding Ki values calculated using the 6D-QSAR model are compared. For Polysantol and Javanol only the Ki values for the energetically most favorable conformation are reported. The IC50 values for the remaining stereoisomers are as follows: 140 ± 173 μm Polysantol (S,S), 216 ± 122 μm Polysantol (S,R), 2.86 ± 4.45 mm Polysantol (R,R), 2.41 ± 2.64 mm Javanol (R,S), 176.7 ± 567 μm Javanol (S,S), and 40.73 ± 23.33 μm Javanol (S,R). The Ki values for the remaining stereoisomeric forms are as follows: 299 μm Polysantol (S,S), 83 μm Polysantol (S,R), 1 mm Polysantol (R,R), 931 μm Javanol (R,S), 68 μm Javanol (S,S), and 15 μm Javanol (S,R).
| Compound | EC50 of ERactivation | Ki of ERcompetitive binding | Ki of ER predicted |
|---|---|---|---|
| Estradiol | 0.16 ± 0.01 nm | 1.6 ± 1 nm | 2 nm |
| MC | 86 ± 8 mm | 76 ± 6 μm | 21 μm |
| Polysantol (R,S)a | 98 ± 23 mm | 56 ± 5 μm | 26 μm |
| Javanol (R,R)a | 50 ± 4 mm | ca. 50 μm | 5 μm |
| Androstenol | 52 ± 3 mm | ca. 25 μm | 10 μm |
a The indicated stereoisomers were used for predicting Ki values, whereas experiments were performed using mixtures of stereoisomers.
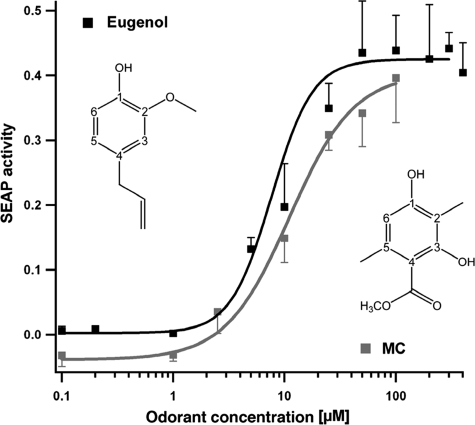
Although molecules of the sandalwood odorant family were shown to elicit signaling responses in a limited number of olfactory neurons isolated from rat (54), no ORs have thus far been cloned and recombinantly expressed that are responsible for the cellular response. Screening an odorant library for compounds that specifically activate mOR-EG in the reporter assay, we discovered that MC is a specific agonist for the mouse eugenol odorant receptor mOR-EG, a receptor described recently by Katada and co-worker (26). We performed functional studies coupling mOR-EG activation to the endogenous cAMP pathway and to the expression of the reporter gene for secreted alkaline phosphatase. Dose-dependent responses of MC in comparison to eugenol indicate that both molecules are similarly potent agonists (Fig. 2); based on our docking simulations and point mutation analysis these molecules bind similarly to the receptor (for details see legend of Fig. 4). Using the same assay the sandalwood odorants of our present study did not elicit any detectable responses on mOR-EG activation. The mOR-EG-specific ligand eugenol, on the other hand, although capable of displacing radioactively labeled E2 from the purified ER, did not show any detectable estrogenic or anti-estrogenic activities in a cellular reporter gene assay (60).
FIGURE 2.
Dose-dependent activation of mOR-EG in HEK293 cells by its cognate ligand eugenol and by the odorant MC. cAMP-dependent cAMP-response element secreted alkaline phosphatase reporter gene activity of mock-transfected cells was subtracted from the secreted alkaline phosphatase activity induced by the corresponding compounds applied.
FIGURE 4.
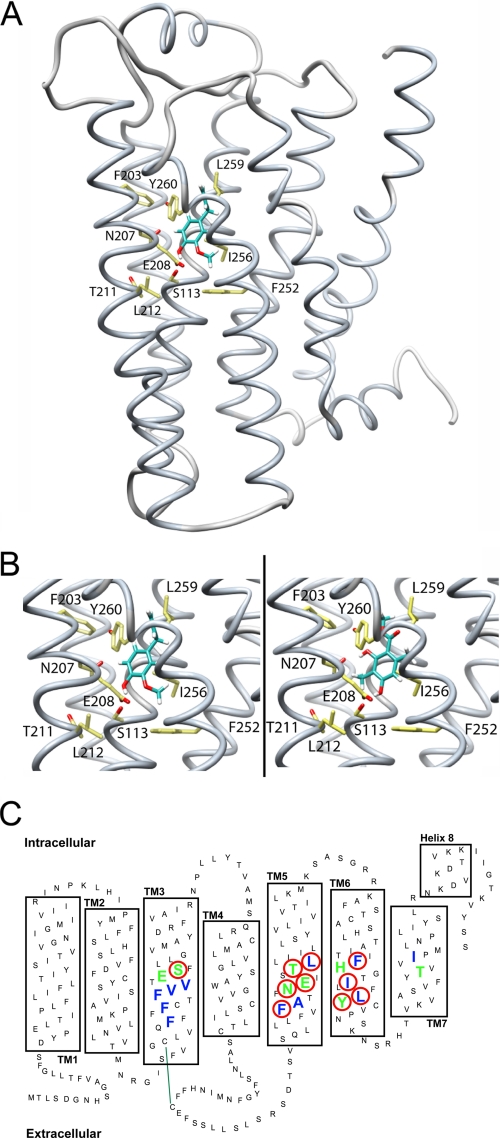
Structural model of the odorant receptor mOR-EG. A, backbone structure of mOR-EG in complex with MC (shown as a ball-and-stick model). B, close-up view of Eugenol (left) and MC (right) docked into the mOR-EG binding site. Site-directed mutagenesis of amino acid residues Ser-113 (S113), Asn-207 (N207), Thr-211 (T211), Leu-212 (L212), Phe-252 (F252), Ile-256 (I265), and Leu-259 (L259) reduce the activity for Eugenol at a ratio of EC50(wt)/EC50(mut) < 0.5 (18). Our own results show that mutants S113A (Eugenol, EC50: 115 ± 20 μm; MC, EC50: 122 ± 20 μm), E208D (Eugenol, EC50: 98 ± 10 μm; MC: no response), F203W (Eugenol, EC50: 96 ± 13 μm; MC: no response), and Y260F (Eugenol, EC50: 103 ± 16 μm; MC, EC50: 120 ± 25 μm) reduce receptor activity for both Eugenol and MC as compared with the wild-type mOR-EG (Eugenol, EC50: 10 ± 1 μm; MC, EC50: 11 ± 2 μm). In our molecular model, both Eugenol and MC form hydrogen bond interactions with the cluster of polar residues around Ser-113, whereas the remaining interactions with the receptor are hydrophobic. C, schematic representation of the receptor binding site: amino acid residues contributing to hydrophobic interactions are blue, and those contributing to polar interactions with MC are green; mutated residues significantly reducing receptor activation are highlighted with a red circle. Green line indicates disulfide bond between conserved cysteins.
Thus far, only odorants structurally related to eugenol, a major odorant in clove oil, have been identified as mOR-EG-specific ligands (26). All these active molecules comprise a benzene ring with substitutions at three fixed positions, including an oxygen at position C1 capable of forming hydrogen bonds and additional variable substitutions at positions C2 and C4 (Fig. 2) involved in multiple hydrophobic contacts with the receptor (26). The penta-substituted benzene ring of MC harboring two hydroxyl groups, two methyl groups, and an ester side chain is therefore quite different from previously described mOR-EG-specific ligands, especially because of the additional functional groups at the C3 and C5 positions (Fig. 2). Our finding indicates that the binding site of mOR-EG accepts variable ligand structures with broader molecular features than previously anticipated (26).
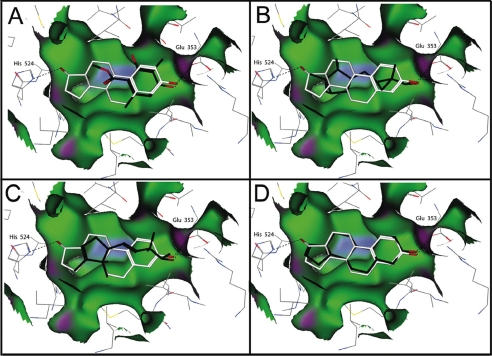
To better understand the molecular interactions of MC with both ER and mOR-EG, we performed molecular modeling of the two structurally diverse receptor·ligand complexes. For ER more than 20 high resolution crystal structures of its ligand binding domain in complex with different agonists or antagonists have been solved (61–63) allowing detailed atomistic simulations of odorant binding. The overall promiscuity of ER can be attributed (i) to its large hydrophobic binding cavity (depicted in green in Fig. 3), which has a probe-accessible volume of 450 Å3 nearly twice of the molecular volume of the natural agonist E2 (245 Å3) and (ii) to the minimal requirement of an effective ligand to contain an aromatic ring, but the remainder of the binding pocket accepting a number of different hydrophobic groups (61–63). The E2·ER complex is characterized by the two residues His-524 and Glu-353 hydrogen-bonded directly to the hormone, and a network of water-mediated hydrogen bonds involving the side chain of Arg-394 and the backbone carbonyl of Phe-404 (61–63). We applied molecular docking simulations to model the interactions of the odorant molecules with the ER (32). Obviously, MC is smaller than E2 and therefore does not exhibit the same tight interactions with ER as does E2. However, the energetically most favorable docked conformation retains one of the key hydrogen bonds to Glu-353 (corresponding to the specific 3-hydroxy interaction in the E2 complex), and the aromatic moiety of MC superposes well with the steroid A ring of E2 mimicking also its hydrophobic contacts to the receptor. Both Javanol and Polysantol are chiral molecules with several stereo-chemical centers. For the docking simulations, all possible stereoisomeric forms, unless known to be inactive, were docked into the receptor. In the docked complex, Javanol, Polysantol, and Androstenol occupy almost the entire length of the binding pocket and can adopt two orientations, where the hydroxyl group of the molecule can form a hydrogen bond with either Glu-353 or His-524 (Fig. 3). Although MC and Javanol exerted quite similar concentration-dependent effects in our ligand-displacement experiments, the two molecules acted differently in the reporter assay. Javanol, which is larger and according to our docking simulation occupies more available space of the binding cavity, is also a stronger ER-specific agonist than MC. Probably Javanol, due to its size and shape, is capable of performing more functional interactions with ER, which favors essential conformational changes that result in receptor activation. All odorant compounds and also androstenol significantly increased the proliferation of MCF7 cells as compared with non-treated control cells. Similarly as found in the reporter gene assay performed in MCF7 the odorant Javanol exhibited the highest activity in the proliferation assay. The ER-mediated proliferative effect of the odorants was reversible by the addition of the antagonist ICI 182,780 (Fig. 1D).
FIGURE 3.
Structural model of human ER with bound ligands. A–D, estradiol (white); A, Mousse Cristal (black); B, Javanol (R,R) (black); C, Polysantol (R,S) (black); and D, Androstenol (black). Hydrogen bonds to residues His-524 and Glu-353 of the ER are shown as dotted lines. Hydrophobic regions of the molecular surface of the receptor are green, mild polar regions are blue, and hydrogen bonding are violet. Chemical structures of ligands are depicted in Fig. 1E.
To estimate the binding affinities of the different ligands to ER, we applied multidimensional quantitative structure activity relationship (6D-QSAR) using up to four energetically favorable conformations for each of the docked odorants. Multidimensional QSAR methods represent each ligand molecule by an ensemble of conformations in a three-dimensional receptor structure representation, allowing for a potentially flexible receptor site (mimicking local induced fit) and combines different solvation models (37). The estimated values of the inhibition constants (Ki, Table 1) for the ER-specific odorants correlate reasonably well with the experimental Ki and EC50 data. They are two to three times lower for MC, Polysantol, and androstenol, and about 10 times lower for Javanol. The differences may arise because we used in the experiments mixtures of stereoisomers of the particular odorants, whereas the in silico calculations were performed with the pure stereoisomers, which exhibited the energetically most favorable conformation in complex with the receptor.
DISCUSSION
Apparently, the ligand binding site of the ER is quite dynamic (64); conformational changes can be induced by small lipophilic ligands such as a phenol derivative, which reacted as an ER-specific agonist favoring the agonist-bound conformation of helix 12 (65). Also parabens, which are similar in size and shape to MC, were shown to exhibit estrogenic activity (66). Furthermore, MC fits well to the binding pocket of ER (Fig. 3) and therefore might act as a partial agonist. In contrast to the ER, the molecular determinants governing the interactions between odorants and their corresponding ORs are less well understood due to lack of experimentally determined high resolution structures and specific binding affinity data for this large family of GPCRs. The binding sites of a few ORs have been modeled on the basis of the known crystallographic structure of rhodopsin (26, 67, 68) or by assembling and optimizing the packing of the seven-helix TM bundle of ORs in lipid bilayer (69). Alternatively, an approach to classify OR binding sites based on sequence comparison has been proposed (70). To better understand the mode of recognition of MC by the mOR-EG, we built structural models for the odorant·receptor complex on the basis of the recently solved high resolution crystallographic structure of β2-adrenergic receptor (38), which together with the ORs and rhodopsin belongs to the class A family of GPCRs (39). Combining our own and previous results (26) of the functional analysis of the mOR-EG binding site involving site-directed point mutations, we established a three-dimensional model of the mOR-EG in complex with eugenol and the MC odorant (Fig. 4).
The binding site of the receptor is formed by residues from transmembrane (TM) helices III, V, and VI and the second extracellular loop. The evolutionary relationship and alignment between TM I to IV and VII of the β2-adrenergic GPCR and mOR-EG receptor can be readily established. However, due to the evolutionary and functional distance between the β2-adrenergic and OR families, the per-residue correspondence in TM V and VI cannot be resolved unambiguously by sequence alignment methods. Therefore, a series of alternative alignments was used to generate an ensemble of comparative receptor models, which were evaluated for consistency with results from mutational analysis studies (see above), Bayesian analysis of mutual information in OR multiple sequence alignments (41), and the molecular docking results. The most favored model based on these criteria shows a mainly hydrophobic binding pocket with a pronounced network of polar interactions by residues Ser-113, Asn-207, Glu-208, Thr-211, and His-253 at the bottom of the cleft (Fig. 4B) connecting TM III, V, and VI. The major differences between the binding sites of our current model and previous results (26) are due to a different target-template alignment of TM V, and intrinsic structural differences between the β2-adrenergic receptor and rhodopsin receptor, in particular, the relative orientation of TM III and V (38).
All active odorants recognized by the mOR-EG have at least one polar group, which in our model is interacting with this polar patch. In particular Ser-113 (26) and Glu-208 seem to define receptor specificity for the polar moiety, whereas most of the remaining MC- and eugenol-receptor interactions are hydrophobic (see Fig. 4B). In analogy to ER, the interaction of MC with mOR-EG involves the formation of specific hydrogen bonds within a mainly hydrophobic binding site. However, the two evolutionary unrelated protein folds give rise to distinct binding pockets with different shapes and character: although the ligand binding site of ER is formed by a large hydrophobic cavity, the ligand binding site of mOR-EG lies at the bottom of a solvent-accessible cleft. GPCRs are predicted to exist in a dynamic equilibrium of agonist and antagonist conformational states, eventually with intermediate states (71). Our current OR model based on β2-adrenergic receptor in complex with the partial inverse agonist carazolol only allows for qualitative interpretations, e.g. in the context of planning further functional site-directed mutagenesis experiments, and studying the structure-activity relationship of mOR-EG-specific ligands with newly discovered odorants structurally different from the originally found cognate ligand eugenol. Quantitative predictions of ligand specificity of ORs remains a goal for the foreseeable future. In contrast, the detailed atomistic model of the ER has turned out to correlate well with the experimental observations and might open the possibility of systematically screening libraries of odorants and other non-volatile small molecules in silico for their potential to activate nuclear hormone receptors. The functional relevance of our present finding that specific odorant molecules have dual activities both on OR and on ER is highlighted by previous results showing the expression of ER in olfactory sensory neurons (72). These neurons respond both to sex steroids and to odorant molecules by secreting the gonadotropin-releasing hormone providing evidence for a direct link between the olfactory and neuroendocrine systems in human (72, 73). A potential functional role of the two receptors in olfactory organogenesis or in the neuroendocrine network that controls human productive behavior is conceivable.
Our approaches are also of general importance for evaluating potential risks of chemical compounds used in fragrances. There is evidence that the human skin does not act as a barrier to certain small, lipophilic compounds (mass < 500 Da) (74). Most odorants including those used in our present study fulfill these criteria and might therefore enter the human body via the skin. Polycyclic musk fragrances, for instance, were shown to accumulate in human adipose tissue and human milk (37). A daily administration of these odorants might therefore result in local concentrations far beyond the limit where endocrine activities become relevant. In addition, estrogenic odorants might act cumulatively on ER activation.
The dual active odorant molecules of the present study exhibit distinct lower estrogenic potency than the natural hormone. However, the physiological relevance of our findings is underlined by the fact that they were achieved on the natural, endogenous ER expressed in human breast cancer cells (MCF7) representing a widely accepted assay system for detecting estrogenic activities (19).
Supplementary Material
Acknowledgments
We are grateful to A. Vendani, M. Smiesko, and M. Spreafico for help and support with the QSAR analysis.
This work was supported by the SystemsX program of the Swiss National Science Foundation and by internal grants of the Ecole Polytechnique Fédérale de Lausaunne.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- OR
- olfactory receptor
- ER
- human estrogen receptor α
- E2
- 17β-estradiol
- DMEM
- Dulbecco's modified Eagle's medium
- YFP
- yellow fluorescent protein
- FCS
- fluorescence correlation spectroscopy
- GPCR
- G-protein-coupled receptor
- TM
- transmembrane
- MC
- Mousse Cristal
- mOR-EQ
- mouse eugenol odorant receptor
- QSAR
- quantitative structure activity relationships.
REFERENCES
- 1.Buck L., Axel R. (1991) Cell 65, 175–187 [DOI] [PubMed] [Google Scholar]
- 2.Mombaerts P. (1999) Science 286, 707–711 [DOI] [PubMed] [Google Scholar]
- 3.Zozulya S., Echeverri F., Nguyen T. (2001) Genome Biology http://genomebiology.com/2001/2/6/RESEARCH/0018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mombaerts P. (2004) Nat. Rev. Neurosci. 5, 263–278 [DOI] [PubMed] [Google Scholar]
- 5.Touhara K., Vosshall L. B. (2009) Annu. Rev. Physiol. 71, 307–332 [DOI] [PubMed] [Google Scholar]
- 6.Shepherd G. M. (2006) Nature 444, 316–321 [DOI] [PubMed] [Google Scholar]
- 7.Kraft P., Bajgrowicz J. A., Denis C., Fráter G. (2000) Angew. Chem. Int. Ed. Engl. 39, 2980–3010 [DOI] [PubMed] [Google Scholar]
- 8.Spehr M., Gisselmann G., Poplawski A., Riffell J. A., Wetzel C. H., Zimmer R. K., Hatt H. (2003) Science 299, 2054–2058 [DOI] [PubMed] [Google Scholar]
- 9.Fukuda N., Yomogida K., Okabe M., Touhara K. (2004) J. Cell Sci. 117, 5835–5845 [DOI] [PubMed] [Google Scholar]
- 10.Feldmesser E., Olender T., Khen M., Yanai I., Ophir R., Lancet D. (2006) BMC Genomics 7, 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neuhaus E. M., Zhang W., Gelis L., Deng Y., Noldus J., Hatt H. (2009) J. Biol. Chem. 284, 16218–16225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pluznick J. L., Zou D. J., Zhang X., Yan Q., Rodriguez-Gil D. J., Eisner C., Wells E., Greer C. A., Wang T., Firestein S., Schnermann J., Caplan M. J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 2059–2064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krautwurst D., Yau K. W., Reed R. R. (1998) Cell 95, 917–926 [DOI] [PubMed] [Google Scholar]
- 14.Jacquier V., Pick H., Vogel H. (2006) J. Neurochem. 97, 537–544 [DOI] [PubMed] [Google Scholar]
- 15.Doszczak L., Kraft P., Weber H. P., Bertermann R., Triller A., Hatt H., Tacke R. (2007) Angew. Chem. Int. Ed. Engl. 46, 3367–3371 [DOI] [PubMed] [Google Scholar]
- 16.Touhara K. (2007) Neurochem. Int. 51, 132–139 [DOI] [PubMed] [Google Scholar]
- 17.Jacquier V., Prummer M., Segura J. M., Pick H., Vogel H. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 14325–14330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lacroix M., Leclercq G. (2004) Breast Cacer Res. Treat. 83, 249–289 [DOI] [PubMed] [Google Scholar]
- 19.Soto A. M., Sonnenschein C., Chung K. L., Fernandez M. F., Olea N., Serrano F. O. (1995) Environ. Health Perspect. 103, 113–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang H., Tong W., Perkins R., Soto A. M., Prechtl N. V., Sheehan D. M. (2000) Environ. Health Perspect. 108, 723–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jankevics H., Prummer M., Izewska P., Pick H., Leufgen K., Vogel H. (2005) Biochemistry 44, 11676–11683 [DOI] [PubMed] [Google Scholar]
- 22.Evans R. M. (1988) Science 204, 889–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Lipzig M. M., ter Laak A. M., Jongejan A., Vermeulen N. P., Wamelink M., Geerke D., Meerman J. H. (2004) J. Med. Chem. 47, 1018–1030 [DOI] [PubMed] [Google Scholar]
- 24.Mueller S. O. (2004) Anal. Bioanal. Chem 378, 582–587 [DOI] [PubMed] [Google Scholar]
- 25.Prossnitz E. R., Arterburn J. B., Smith H. O., Oprea T. I., Sklar L. A., Hathaway H. J. (2008) Annu. Rev. Physiol. 70, 165–190 [DOI] [PubMed] [Google Scholar]
- 26.Katada S., Hirokawa T., Oka Y., Suwa M., Touhara K. (2005) J. Neurosci. 25, 1806–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saito H., Kubota M., Roberts R. W., Chi Q., Matsunami H. (2004) Cell 119, 679–691 [DOI] [PubMed] [Google Scholar]
- 28.Weiner S. J., Kollmann P. A., Case D. A., Sing U. C., Ghio C., Alagona G., Profeta S., Weiner P. (1984) 106, 765–784 [Google Scholar]
- 29.Mohamadi F., Richards M. G., Guida W. C., Liskamp R., Lipton M., Caufield C., Chang G., Hendrickson T., Still W. C. (1990) J. Comput. Chem. 11, 440–467 [Google Scholar]
- 30.Cramer C. J., Truhlar D. G. (1992) J. Comput. Aided Mol. Des. 6, 629–666 [DOI] [PubMed] [Google Scholar]
- 31.Shiau A. K., Barstad D., Loria P. M., Cheng L., Kushner P. J., Agard D. A., Greene G. L. (1998) Cell 95, 927–937 [DOI] [PubMed] [Google Scholar]
- 32.Lill M. A., Dobler M., Vedani A. (2006) Chem. Med. Chem. 1, 73–81 [DOI] [PubMed] [Google Scholar]
- 33.Vedani A., Dobler M., Lill M. A. (2005) J. Med. Chem. 48, 3700–3703 [DOI] [PubMed] [Google Scholar]
- 34.Cheng P., Prusoff W. H. (1973) Biochem. Pharmacol. 22, 3099–3108 [DOI] [PubMed] [Google Scholar]
- 35.Reese J. C., Katzenellenbogen B. S. (1992) J. Biol. Chem. 267, 9868–9873 [PubMed] [Google Scholar]
- 36.Blair R. M., Fang H., Branham W. S., Hass B. S., Dial S. L., Moland C. L., Tong W., Shi L., Perkins R., Sheehan D. M. (2000) Toxicol. Sci. 54, 138–153 [DOI] [PubMed] [Google Scholar]
- 37.Rimkus G. G., Wolf M. (1996) Chemosphere 33, 2033–2043 [DOI] [PubMed] [Google Scholar]
- 38.Cherezov V., Rosenbaum D. M., Hanson M. A., Rasmussen S. G., Thian F. S., Kobilka T. S., Choi H. J., Kuhn P., Weis W. I., Kobilka B. K., Sevens R. C. (2007) Science 318, 1258–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palczewski K., Kumasaka T., Hori T., Behnke C. A., Motoshima H., Fox B. A., Le Trong I., Teller D. C., Okada T., Stenkamp R. E., Yamamoto M., Miyano M. (2000) Science 289, 739–745 [DOI] [PubMed] [Google Scholar]
- 40.Armougom F., Moretti S., Poirot O., Audic S., Dumas P., Schaeli B., Keduas V., Notredame C. (2006) Nucleic Acids Res. 34, W604–W608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burger L., van Nimwegen E. (2008) Mol. Syst. Biol. 4, 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bissantz C., Logean A., Rognan D. (2004) J. Chem. Inf. Comput. Sci. 44, 1162–1176 [DOI] [PubMed] [Google Scholar]
- 43.Sali A., Blundell T. L. (1993) J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 44.Sherman W., Day T., Jacobson M. P., Friesner R. A., Farid R. (2006) J. Med. Chem. 49, 534–553 [DOI] [PubMed] [Google Scholar]
- 45.Htun H., Holth L. T., Walker D., Davie J. R., Hager G. L. (1999) Mol. Biol. Cell 10, 471–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stenoien D. L., Nye A. C., Mancini M. G., Patel K., Dutertre M., O‘Malley B. W., Smith C. L., Belmont A. S., Mancini M. A. (2001) Mol. Cell. Biol. 21, 4404–4412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pick H., Jankevics H., Vogel H. (2007) J. Mol. Biol. 374, 1213–1223 [DOI] [PubMed] [Google Scholar]
- 48.Matsuda K., Nishi M., Takaya H., Kaku N., Kawata M. (2008) J. Cell. Biochem. 103, 136–148 [DOI] [PubMed] [Google Scholar]
- 49.Wachsmuth M., Waldeck W., Langowski J. (2000) J. Mol. Biol. 298, 677–689 [DOI] [PubMed] [Google Scholar]
- 50.Gronemeyer H., Gustafsson J. A., Laudet V. (2004) Nat. Rev. Drug Discov. 3, 950–964 [DOI] [PubMed] [Google Scholar]
- 51.Feige J. N., Gelman L., Tudor C., Engelborghs Y., Wahli W., Desvergne B. (2005) J. Biol. Chem. 280, 17880–17890 [DOI] [PubMed] [Google Scholar]
- 52.Deroo B. J., Korach K. S. (2006) J. Clin. Invest. 116, 561–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bain D. L., Heneghan A. F., Connaghan-Jones K. D., Miura M. T. (2007) Annu. Rev. Physiol. 69, 201–220 [DOI] [PubMed] [Google Scholar]
- 54.Bieri S., Monastyrskaia K., Schilling B. (2004) Chem. Senses 29, 483–487 [DOI] [PubMed] [Google Scholar]
- 55.Kaminski R. M., Marini H., Ortinski P. I., Vicini S., Rogawski M. A. (2006) J. Pharmacol. Exp. Ther. 317, 694–703 [DOI] [PubMed] [Google Scholar]
- 56.Schreurs R. H., Quaedackers M. E., Seinen W., van der Burg B. (2002) Toxicol. Appl. Pharmacol. 183, 1–9 [DOI] [PubMed] [Google Scholar]
- 57.Likhite V. S., Stossi F., Kim K., Katzenellenbogen B. S., Katzenellenbogen J. A. (2006) Mol. Endocrinol. 20, 3120–3132 [DOI] [PubMed] [Google Scholar]
- 58.Meegan M. J., Barrett I., Zimmermann J., Knox A. J., Zisterer D. M., Lloyd D. G. (2007) J. Enzyme Inhib. Med. Chem. 22, 655–666 [DOI] [PubMed] [Google Scholar]
- 59.Abdelrahim M., Ariazi E., Kim K., Khan S., Barhoumi R., Burghardt R., Liu S., Hill D., Finnell R., Wlodarczyk B., Jordan V. C., Safe S. (2006) Cancer Res. 66, 2459–2467 [DOI] [PubMed] [Google Scholar]
- 60.Howes M. J., Houghton P. J., Barlow D. J., Pocock V. J., Milligan S. R. (2002) J. Pharm. Pharmacol. 54, 1521–1528 [DOI] [PubMed] [Google Scholar]
- 61.Brzozowski A. M., Pike A. C., Dauter Z., Hubbard R. E., Bonn T., Engström O., Oehman L., Greene G. L., Gustafsson J. A., Carlquist M. (1997) Nature 389, 753–758 [DOI] [PubMed] [Google Scholar]
- 62.Anstead G. M., Carlson K. E., Katzenellenbogen J. A. (1997) Steroids 62, 268–303 [DOI] [PubMed] [Google Scholar]
- 63.Tanenbaum D. M., Wang Y., Williams S. P., Sigler P. B. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 5998–6003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shiau A. K., Barstad D., Radek J. T., Meyers M. J., Nettles K. W., Katzenellenbogen B. S., Katzenellenbogen J. A., Agard D. A., Greene G. L. (2002) Nat. Struct. Biol. 9, 359–364 [DOI] [PubMed] [Google Scholar]
- 65.Yamasaki K., Takeyoshi M., Yakabe Y., Sawaki M., Takatsuki M. (2003) Toxicol. Lett. 142, 119–131 [DOI] [PubMed] [Google Scholar]
- 66.Terasaki M., Kamata R., Shiraishi F., Makino M. (2009) Environ. Toxicol. Chem. 28, 204–208 [DOI] [PubMed] [Google Scholar]
- 67.Schmiedeberg K., Shirokova E., Weber H. P., Schilling B., Meyerhof W., Krautwurst D. (2007) J. Struct. Biol. 159, 400–412 [DOI] [PubMed] [Google Scholar]
- 68.Abaffy T., Malhotra A., Luetje C. W. (2007) J. Biol. Chem. 282, 1216–1224 [DOI] [PubMed] [Google Scholar]
- 69.Floriano W. B., Vaidehi N., Goddard W. A., 3rd (2004) Chem. Senses 29, 269–290 [DOI] [PubMed] [Google Scholar]
- 70.Man O., Gilad Y., Lancet D. (2004) Protein Sci. 13, 240–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Serrano-Vega M. J., Magnani F., Shibata Y., Tate C. G. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 877–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Barni T., Maggi M., Fantoni G., Granchi S., Mancina R., Gulisano M., Marra F., Macorsini E., Luconi M., Rotella C., Serio M., Balboni G. C., Vannelli G. B. (1999) J. Clin. Endocrinol. Metab. 84, 4266–4273 [DOI] [PubMed] [Google Scholar]
- 73.Romanelli R. G., Barni T., Maggi M., Luconi M., Failli P., Pezzatini A., Pelo E., Torricelli F., Crescioli C., Ferruzzi P., Salerno R., Marini M., Rotella C. M., Vannelli G. B. (2004) J. Biol. Chem. 279, 117–126 [DOI] [PubMed] [Google Scholar]
- 74.Brown M. B., Martin G. P., Jones S. A., Akomeah F. K. (2006) Drug Deliv. 13, 175–187 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.