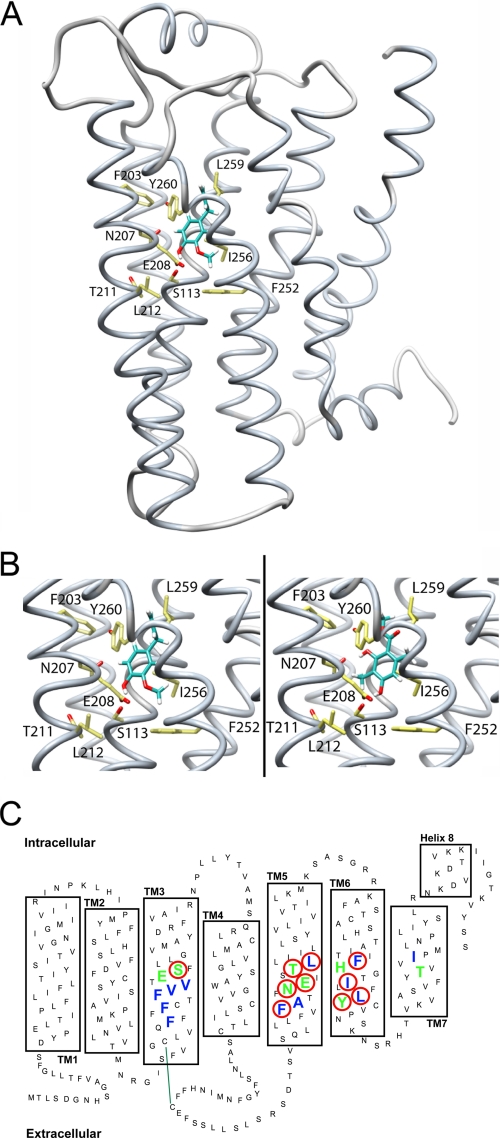
FIGURE 4.
Structural model of the odorant receptor mOR-EG. A, backbone structure of mOR-EG in complex with MC (shown as a ball-and-stick model). B, close-up view of Eugenol (left) and MC (right) docked into the mOR-EG binding site. Site-directed mutagenesis of amino acid residues Ser-113 (S113), Asn-207 (N207), Thr-211 (T211), Leu-212 (L212), Phe-252 (F252), Ile-256 (I265), and Leu-259 (L259) reduce the activity for Eugenol at a ratio of EC50(wt)/EC50(mut) < 0.5 (18). Our own results show that mutants S113A (Eugenol, EC50: 115 ± 20 μm; MC, EC50: 122 ± 20 μm), E208D (Eugenol, EC50: 98 ± 10 μm; MC: no response), F203W (Eugenol, EC50: 96 ± 13 μm; MC: no response), and Y260F (Eugenol, EC50: 103 ± 16 μm; MC, EC50: 120 ± 25 μm) reduce receptor activity for both Eugenol and MC as compared with the wild-type mOR-EG (Eugenol, EC50: 10 ± 1 μm; MC, EC50: 11 ± 2 μm). In our molecular model, both Eugenol and MC form hydrogen bond interactions with the cluster of polar residues around Ser-113, whereas the remaining interactions with the receptor are hydrophobic. C, schematic representation of the receptor binding site: amino acid residues contributing to hydrophobic interactions are blue, and those contributing to polar interactions with MC are green; mutated residues significantly reducing receptor activation are highlighted with a red circle. Green line indicates disulfide bond between conserved cysteins.