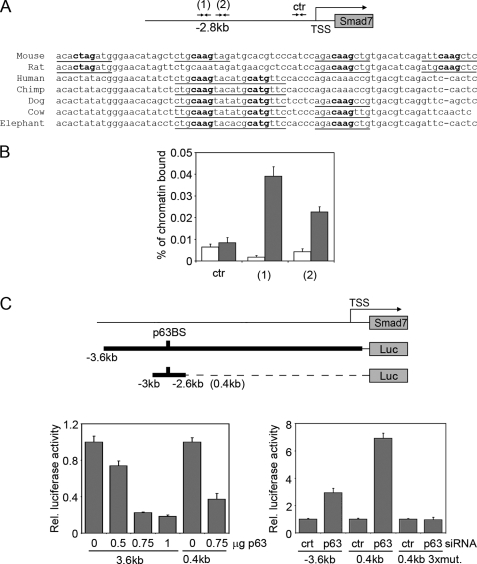
FIGURE 3.
Smad7 is a transcriptional target of p63. A, a conserved p63-binding site is located at −2.8 kb from the Smad7 transcription start site. The predicted p63-binding hemisites are indicated with their nucleotide sequence and phylogenetic conservation in multiple species. Bold nucleotides correspond to the core nucleotide sequence required for p63 binding, whereas underlined nucleotides are matches in the consensus (22, 53). B, specific binding of endogenous p63 to the mouse Smad7 promoter. Primary keratinocytes were processed for ChIP with antibodies specific for p63 (gray bars), or unrelated anti-ERK antibodies as control (white bars), followed by real-time PCR amplification using oligonucleotide primers designed at the indicated position from the transcription start site. The amount of precipitated DNA was calculated relative to the total input chromatin, and expressed as the percentage of the total DNA, as previously described (54). C, the activity of a 3.6-kb Smad7 promoter region and its 0.4-kb fragment spanning the p63-binding sites (p63BS) were measured in the presence of the indicated amounts of a ΔNp63α-expressing construct (p63) (left panel), or p63 or control (ctr) siRNA (right panel). A 0.4-kb fragment containing 2-bp mutations in each of the first three binding hemisites indicated in A (0.4kb 3xmut) was also co-transfected with p63 or control siRNA. Values are expressed as described in Fig. 1A.