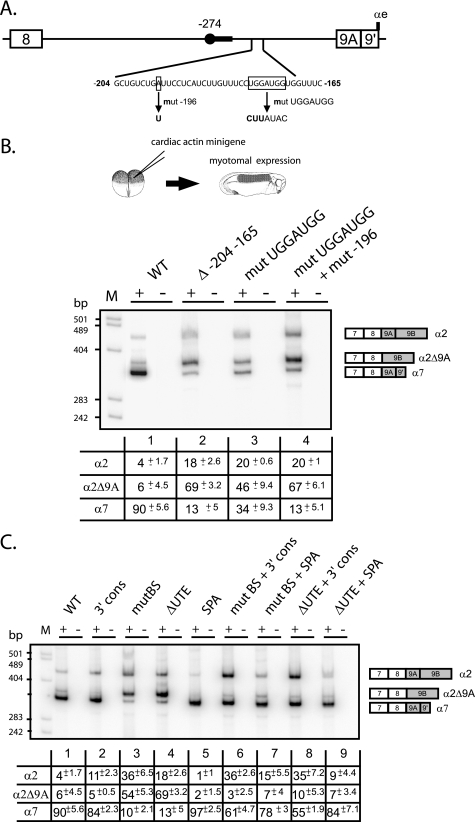
FIGURE 2.
An intronic sequence upstream of exon 9A9′ contributes to its splicing as a 3′-terminal exon in muscle cells. A, schematic map of the intronic region upstream of exon 9A9′. The distant −274 branch site and its associated polypyrimidine tract are represented by a black circle and a black rectangle, respectively. The intronic sequence from −204 to −165 upstream of exon 9A9′ is shown. The generated mutations are shown below the wild-type sequence. B, the wild-type (WT) and the indicated mutant minigenes under the control of the cardiac actin promoter were injected into Xenopus embryos. The mRNA splicing pattern was assayed by 3′-RACE-PCR using a 32P-oligonucleotide specific to the cardiac actin minigene constructs in the presence (+) or absence (−) of reverse transcriptase. The structure of the amplified fragments is indicated on the right. Quantification was performed on a PhosphorImager to yield the proportion of each mRNA. The percentage of each mRNA is given in the table below a representative experiment as mean ± S.D. (n = 20 for the wild type construct and n = 3 for the mutants). C, the distant −274 branch site and UTE are functionally coupled. The Δ−204−165 mutation (ΔUTE) and the mutation of the distant nucleotide −274 branch site (mutBS) were associated with the 3′ cons mutant in which seven consecutive pyrimidine near the dinucleotide AG were introduced or to the SPA mutant in which a strong synthetic poly(A) site was substituted to the αe poly(A) site. The wild type and the indicated mutant minigenes under the control of the cardiac actin promoter were injected into Xenopus embryos. Samples were processed as for B. The percentage of each mRNA is given in the table below a representative experiment as mean ± S.D. (n = 20 for the wild-type construct, and n = 3 for the mutants).