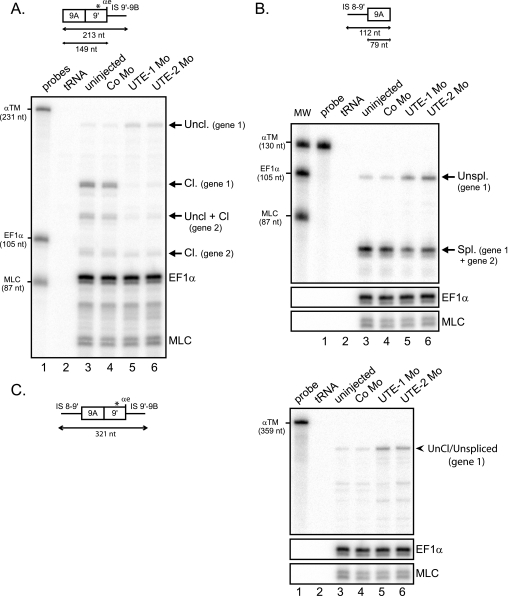
FIGURE 5.
UTE activates the splicing and cleavage/polyadenylation of exon 9A9′. RNase protection analysis of RNA extracted from uninjected embryos or embryos injected with UTE-1 Mo, UTE-2 Mo, or control morpholino (Co Mo). The RNase protection probes are presented above. A, UTE activates the cleavage/polyadenylation of exon 9A9′. RNase protection was realized with an RNA probe extending beyond the αe poly(A) site, as presented at the top. The three-nucleotide difference within exon 9′ between the two α-TM genes is indicated by a star. The identity of uncleaved (Uncl.) and cleaved (Cl.) protected fragments is indicated on the right. B, UTE activates the splicing of exon 9A9′. RNase protection was realized with an RNA probe that covered the 3′ splice site of exon 9A9′, as presented at the top. The identity of unspliced and spliced fragments is shown on the right. C, unprocessed RNAs accumulate in UTE Mo-injected embryos. RNase protection was realized with a probe spanning the 3′ splice site and extending beyond the cleavage site of exon 9A9′, as shown on the left. The protected fragment corresponding to uncleaved/unspliced (Uncl./Unspl.) molecules is indicated on the right. A–C, the protected fragments were quantified by PhosphorImager analysis and normalized to uridine content and EF1-α. The α-TM, EF1-α, and MLC probes were used as molecular weight markers (MW).