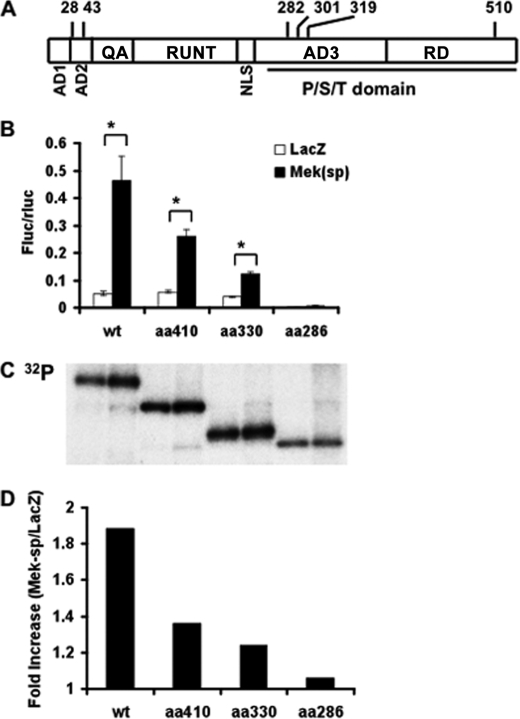
FIGURE 1.
Identification of a region in Runx2 necessary for ERK/MAPK-dependent transcriptional activation and phosphorylation. A, schematic of the domain structure of Runx2 with relevant serine residues indicated. AD1–3, transcriptional activation domains; QA, glutamine/alanine-rich domain; RUNT, runt/DNA-binding domain; NLS, nuclear localization sequence; P/S/T domain, proline/serine/threonine-rich domain; RD, repressor domain (from Ref. 30). B, MAPK-dependent transcriptional activity. COS7 cells were transfected with wild type (WT) Runx2 or the indicated C-terminal deletions in the presence of control (LacZ) or Mek(sp) expression vectors and a 6OSE2-luc reporter as described under “Experimental Procedures.” Firefly luciferase activity was normalized for transfection efficiency using a R. reinformis luciferase plasmid. Asterisk, significantly different from corresponding control, p < 0.01; brackets indicate comparisons made; error bars, ±S.D. C and D, Runx2 phosphorylation. COS7 cell cultures treated as in B were metabolically labeled with [32P]orthophosphate or Tran35S-label as described under “Experimental Procedures.” Runx2 was immunoprecipitated (C) and 32P incorporation was normalized to total 35S-labeled protein in each group and expressed as fold-increase with Mek(sp) stimulation (D).