Abstract
With the goal of identifying hitherto unknown surface exosites of streptokinase involved in substrate human plasminogen recognition and catalytic turnover, synthetic peptides encompassing the 170 loop (CQFTPLNPDDDFRPGLKDTKLLC) in the β-domain were tested for selective inhibition of substrate human plasminogen activation by the streptokinase-plasmin activator complex. Although a disulfide-constrained peptide exhibited strong inhibition, a linear peptide with the same sequence, or a disulfide-constrained variant with a single lysine to alanine mutation showed significantly reduced capabilities of inhibition. Alanine-scanning mutagenesis of the 170 loop of the β-domain of streptokinase was then performed to elucidate its importance in streptokinase-mediated plasminogen activation. Some of the 170 loop mutants showed a remarkable decline in kcat without any alteration in apparent substrate affinity (Km) as compared with wild-type streptokinase and identified the importance of Lys180 as well as Pro177 in the functioning of this loop. Remarkably, these mutants were able to generate amidolytic activity and non-proteolytic activation in “partner” plasminogen as wild-type streptokinase. Moreover, cofactor activities of the 170 loop mutants, pre-complexed with plasmin, against microplasminogen as the substrate showed a similar pattern of decline in kcat as that observed in the case of full-length plasminogen, with no concomitant change in Km. These results strongly suggest that the 170 loop of the β-domain of streptokinase is important for catalysis by the streptokinase-plasmin(ogen) activator complex, particularly in catalytic processing/turnover of substrate, although it does not seem to contribute significantly toward enzyme-substrate affinity per se.
Introduction
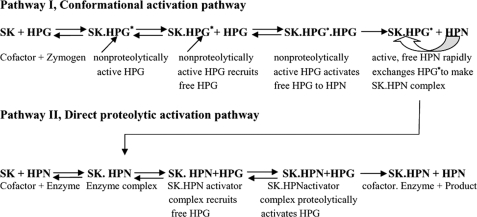
Plasminogen activation, a key event in physiological fibrinolysis, is a widely employed therapeutic means to rapidly alleviate circulatory disorders that arise due to obstruction of blood flow by pathological thrombi (1). Streptokinase (SK),2 a bacteria-derived thrombolytic protein, activates plasminogen by forming a high affinity stoichiometric complex with “partner” plasminogen, which, after a conformational activation step, acquires the capability to selectively cleave the Arg561-Val562 scissile peptide bond in “substrate” plasminogen, thereby converting the latter into HPN, plasmin (2, 3). Elegant equilibrium binding and kinetic studies directed toward resolving the steps involved in the phenomenon of conformational activation of HPG by SK demonstrate a relatively rapid formation of a SK·HPG* complex, a “conformationally activated” state, due to the generation of a nonproteolytically activated active center, which is also amidolytically active (4). This conformationally activated SK·HPG* complex can then recruit a substrate molecule of HPG (3, 9) and proteolytically activate it, likely through inter-molecular proteolytic scission, into HPN. In addition, the HPG moiety in the SK·HPG* activator complex can exchange with free HPN preferentially (because the latter has been shown to have nearly three orders higher affinity for SK) to form a SK·HPN complex (the fully mature activator complex) possessing strong HPG activation capability (5, 7) (Fig. 1). One of the fascinating aspects of HPG activation reaction is the molecular mechanisms whereby the binding of SK to HPN modulates the substrate specificity of the latter (with, essentially, a trypsin-like broad specificity), virtually re-programming it to a very high degree of preference for the Arg561-Val562, the scissile peptide bond of substrate HPG that leads to the HPG proteolytic activation (5–8). The crystal structure of SK complexed with μPN (the isolated catalytic domain of HPN), with which SK forms a tight binary complex as it does with full-length HPG, reveals that the domains of SK form a “three-sided crater,” with the μPN situated at the bottom of a valley into which the substrate's catalytic domain, bearing the “target” scissile peptide bond to be cleaved, can potentially dock (9). Besides the crystal structure, the crucial role of long range protein-protein interactions between the enzyme (SK·plasmin) and its macromolecular substrate HPG have become increasingly clear through domain truncation studies on both SK and HPG, and various other approaches (10–14). Interestingly, the crystal structure of SK·μPN reveals that each of the three SK domains contains a number of both defined and undefined loops, some of which are positioned to potentially bind with docked substrate plasminogen (9). An epitope in SK, the so-called 88–97 loop of streptokinase α-domain, has been identified recently (12) as being important in both 1:1 as well as ternary, enzyme-substrate interactions. However, its overall contribution toward plasminogen activation cannot be described as major, because its mutation was found to abolish only a relatively small fraction of the overall activity of SK·HPN against HPG and μPG. In addition to the 88–97 loop, the 250 loop, which is prominently located in a surface-accessible region of the β-domain so as to potentially interact with an incoming substrate molecule (14), has also been implicated in the interaction of the SK·HPN activator complex with the kringles of substrate HPG, particularly in terms of enhancing enzyme-substrate affinity (14). Mutations in the 250 loop, especially the two lysine residues at its tip, have been demonstrated to result in a 6- to 7-fold decrease in the affinity of the SK·HPN complex with the substrate by surface plasmon resonance as well as steady-state kinetics, with little change in the kcat for HPG activation (14). This loop has been shown to enhance the affinity of SK·HPG* and SK·HPN catalytic complexes toward substrate plasminogen through kringle-mediated interactions (14), via the fifth kringle (15). It is notable, however, especially judging from the quantitative catalytic contributions that both these substrate interacting sites/exosites make toward catalysis (12, 14, 15), that these are probably not sufficient in themselves to fully account for the observed catalytic activity of the SK·HPN complex. This observation prompted us to look for other potential exosites, particularly among the surface-located loops, that could be important in the action of SK on human plasminogen.
FIGURE 1.
Currently understood mechanism of plasminogen activation by streptokinase. SK forms an equimolar complex with HPG (Pathway I) that leads to the generation of an active center in the complex (SK·HPG*) without cleavage of scissile peptide bond in the partner HPG. This “virgin complex” then recruits free HPG as substrate and converts the latter into HPN. The HPN so formed rapidly exchanges with the HPG* from SK·HPG* complex due to its multifold higher affinity for SK in comparison to HPG. Alternatively, the SK can directly combine with the HPN to make SK·HPN activator complex (Pathway II). The activator complex then catalytically acts on substrate HPG molecules and converts them into HPN.
The location of the 170 loop in the crystal structure of SK-μPG (9, 16) suggests that this loop does not likely participate in the formation of the binary complex, which hints at its possible importance in interaction with substrate plasminogen. The present study, indeed, demonstrates for the first time, a direct involvement of this loop in SK-mediated catalysis through (i) competitive assays using synthetic peptides derived from the loop sequence and (ii) alanine-scanning mutagenesis of the various residues in the 170 loop. The results, presented below, reveal that this loop contributes in a major way toward catalytic processing of the substrate plasminogen, because various point-mutations within this loop resulted in more than an order of drop in kcat even though substrate affinity of the mutants, as well as their capability to engender the non-proteolytic activation in partner plasminogen, as tested through the activation of scissile-peptide bond mutant of microplasminogen (μPG R561A) remains unaltered. Thus, the present study identifies a new exosite in SK that is important in the activation of HPG.
EXPERIMENTAL PROCEDURES
Materials
Glu-plasminogen was either purchased from Roche Diagnostics GmBH, Germany, or purified from human plasma by affinity chromatography (17). The T7 RNA polymerase-promoter-based expression vector, pET 23(d), and Escherichia coli strain BL21 (DE3) were products of Novagen Inc. (Madison, WI). Thermostable DNA polymerase (pfu TurboTM) was obtained from Stratagene Inc. (La Jolla, CA). Oligonucleotide primers were supplied by Biobasic Inc., Canada. Phenyl-agarose 6XL was procured from Prometic Biosciences Ltd., UK, and DEAE-Sepharose (fast-flow) from Amersham Biosciences. Urokinase, ϵ-aminocaproic acid, sodium cyanoborohydride, and l-lysine were purchased from Sigma. All other reagents used were of the highest analytical grade available.
Peptide Synthesis
Custom synthesis of the peptides derived from the 170 loop primary structure, namely CQFTPLNPDDDFRPGLKDTKLLC, or its mutated sequence, namely CQFTPLNPDDDFRPGLADTKLLC, was carried out using standard Fmoc (N-(9-fluorenyl)methoxycarbonyl)-based solid-phase peptide chemistry on an automated peptide synthesizer model PS3 by M/s USV, Mumbai, India. After synthesis and cleavage by standard procedures, the peptides were purified on reverse transcription-high-performance liquid chromatography and disulfide-constrained peptide, were cyclized by H2O2 treatment. Linear peptides were N-terminally acetylated and their C-terminal amidated.
Design and Construction of Various SK Loop Mutants
The SK gene from Streptococcus equisimilis H46A strain, a type C streptococcus, was earlier cloned in pET 23d vector (11), and this construct was used to prepare all substitution mutants. The substitution point-mutations of the 170 loop of the β-domain of SK were prepared by using a QuikChange® mutagenesis kit obtained from Stratagene Inc., which involves usage of two complementary primers having the desired mutation at the center (see supplemental Table SI for a list of the primers designed for the various mutants).
Expression and Purification of wtSK/SK Loop Mutants
The wtSK and 170 loop mutants cloned in pET 23d were expressed intracellularly in E. coli BL21 (DE3) cells as inclusion bodies under the control of the T7 phage RNA polymerase promoter after induction with isopropyl-1-thio-β-d-galactopyranoside, and purified as described (11).
Preparation of HPN
Plasmin (HPN), the active form of HPG, was prepared by digesting Glu-HPG with urokinase covalently immobilized on agarose beads using a ratio of 300 Plough units/mg HPG in 50 mm Tris-Cl, pH 8.0, 25% glycerol, and 25 mm l-lysine at 22 °C for 10 h (11).
Preparation of μPG
μPG, the catalytic domain of plasminogen (residues Lys530–Asn790) made devoid of all five kringles, was prepared by cleavage of HPG by HPN under alkaline conditions (0.1 n glycine/NaOH buffer, pH 10.5) at 25 °C (18). Recombinant μPG and its scissile peptide-bond mutant, R561A, were expressed and purified as described earlier (19).
Assays for Determining Inhibition of HPG Activator Activity of Pre-formed wtSK·HPN or Mutant SK·HPN Complex by Peptides Comprising 170 Loop Amino Acid Sequence/Mutated 170 Loop Sequence
The SK·HPN activator complex was formed by preincubating equimolar concentrations (0.5 μm each) of SK and HPN in 50 mm Tris-Cl, pH 7.5, and 0.5% bovine serum albumin at 22 °C for 1 min (20). This enzyme complex, at a final concentration of 5 nm, was added to reaction mixes already containing various concentrations of individual peptides (0–500 μm) in assay buffer containing 50 mm Tris-Cl, pH 7.5, and 100 mm NaCl preincubated with 0.05 μm of HPG for 10 min. The generation of plasminogen activator activity was measured spectrophotometrically at 405 nm by recording the increase in rate of release of p-nitroanilide from the chromogenic substrate by the human plasmin generated as a result of HPG activation for a period of 10 min at 22 °C. A control reaction contained all components except the test peptide. The specific activity at each concentration of inhibitory peptide was calculated by obtaining the slopes of the activation progress curves as change in absorbance/t2. The extent of inhibition by different concentrations of the given peptide was determined and expressed in percentage relative to control reactions (taken as 100%) conducted in the absence of peptide.
Assays for Studying the Activation of HPG by wtSK and SK Mutants
A one-stage colorimetric assay method, was used to measure the kinetics of HPG activation by wtSK or its mutants (21). The change in absorbance at 405 nm was then measured as a function of time (t) in a Molecular Devices Versamax microplate reader at 22 °C. The activator activities were obtained from the slopes of the activation progress curves as change in absorbance/t2 (21).
N-terminal Methionine Removal from wtSK/170 Loop Mutants
The N-terminal methionine was removed from wtSK and 170 loop mutants using methionine aminopeptidase (pfu MetAP) enzyme by a procedure, as described previously (22). The extent of removal of the N-terminal methionine was determined by N-terminal sequence analysis on Applied Biosystems sequencer, Model 491A, and also through quantitative amino acid composition analysis.
Esterolytic Activation of Equimolar HPG·SK/170 Loop Mutant Complex
To monitor the active site formation 7 μm HPG was added to an assay cuvette containing 7.5 μm wtSK/mutant, 100 μm NPGB, and 10 mm sodium phosphate buffer, pH 7.5. The “burst” of p-nitrophenol release due to acylation of active center was monitored at 410 nm as a function of time at 22 °C (2, 23, 24).
Amidolytic Activation by Equimolar wtSK/SK Loop Mutant of R561A μPG Mutant
R561A μPG mutant (final concentration, 5.0 μm) was incubated with 5.5 μm wtSK or a mutant SK, in 0.5% bovine serum albumin, 50 mm Tris-Cl buffer, pH 7.5. The time course of amidolytic activity generation was determined after transferring an aliquot of the complex to a 100-μl reaction well containing assay buffer, 50 mm Tris-Cl, pH 7.5, 100 mm NaCl along with 1 mm Chromozym®PL (the transfer time (including initial complexation was ∼30 s). Final concentration of the complex in the amidolytic assay reaction was 10 nm. The change in absorbance at 405 nm was monitored spectrophotometrically as a function of time (19).
Determination of Steady-state Kinetic Constants for HPG/μPG Activator Activity of wtSK/SK Loop Mutants
The kinetics of HPG/μPG activation by HPN·wtSK/SK-loop mutant complexes was measured by transferring suitable aliquots (usually, so as to attain a 0.5 nm final concentration in the assay mix) of preformed HPN·wtSK/SK loop mutant complexes to the reaction wells containing varying concentrations of substrate HPG/μPG in assay buffer (50 mm Tris-Cl buffer, pH 7.5, and 100 mm NaCl) also containing 0.5 mm chromogenic substrate Chromozym®PL. The generation of activator activity was monitored at 22 °C at 405 nm as before. The kinetic parameters for HPG/μPG activation were then calculated from Michaelis-Menten (v versus S) and inverse (1/v versus 1/S) Lineweaver-Burk plots (20).
Kinetic Analysis of Protein-Protein Interactions by Surface Plasmon Resonance-Ternary Interaction Analysis
A surface plasmon resonance-based biosensor was also used to measure the rate and equilibrium dissociation constants describing interactions between soluble analytes (HPG and μPG) and wtSK/SK loop mutants complexed with immobilized HPG, a situation simulating substrate binding to binary complex and hereafter referred to as ternary interaction as described previously (14). Experiments were performed at 25 °C in HBS “running buffer” (30 mm Hepes, pH 7.4, 135 mm NaCl, and 1 mm EDTA) supplemented with 0.05% Tween 20 and 5 mm NPGB. The NPGB was included to prevent plasmin-mediated proteolysis (14) (see supplemental Figs. S4 and S5 for binding sensorgrams). After subtracting the nonspecific response the association rate constants (kon values), the dissociation rate constants (koff values) and the equilibrium binding constant (KD) were calculated from sensorgrams by non-linear fitting of the association and dissociation curves according to a 1:1 binding model, using BIACORE 3000 evaluation software.
RESULTS AND DISCUSSION
Selective Inhibition of the Activator Activity of the Preformed wtSK·HPN Enzyme Complex by Peptides Comprising Residues 165–185 of the Primary Structure of SK
With the objective to explore the possible role of the 170 loop of SK in plasminogen activation, a set of peptides was synthesized corresponding to residues Pro165 to Leu185 (each containing 23 residues), including unconstrained, linear, and peptides constrained at the N and C termini through an intramolecular disulfide bond (25) after cysteine additions at both ends (see “Experimental Procedures” for details). The synthetic peptides were then examined for their ability to selectively inhibit activation of sub-saturating concentrations of substrate HPG by the preformed wtSK·HPN enzyme complex. Such a peptide mimotope approach is closely similar to our earlier studies to map another important substrate-interacting site in SK (26, 27).
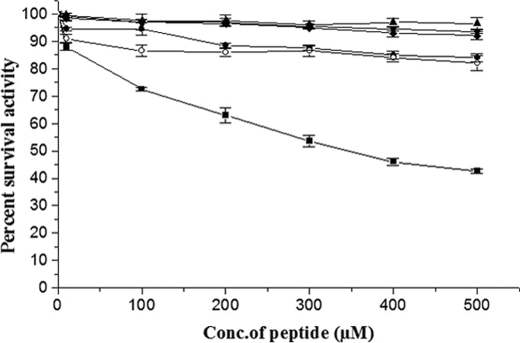
Remarkably, Peptide 2 (wild-type 170 loop sequence constrained through a disulfide bond) inhibited the activation of substrate HPG in a concentration-dependent manner in the low micromolar range of concentration, whereas the linear peptide, i.e. Peptide 1, could not inhibit substrate HPG activation significantly compared with uninhibited controls (see Fig. 2 and supplemental Fig. S1 for actual progress curves showing direct inhibition by the constrained peptide). This observation indicated that the linear, 170 loop peptide probably remains in an extended conformation incapable of competing with SK·HPN for its substrate interactions, whereas the cyclic peptide adopts a conformation that mimics the 170 loop epitope in the native structure of SK. Apart from testing for inhibition of the preformed wtSK·HPN enzyme complex, the two peptides were tested for their ability to inhibit amidolytic activity of HPN, on the one hand, and also the amidolytic activity of SK·HPN complex, on the other. In both cases, no observable inhibition could be seen in the amidolytic activity of free HPN as well as wtSK·HPN complex, which suggests that this 170 loop peptide selectively interferes in the interaction of the activator enzyme with substrate HPG, and thereby competitively inhibits its activation.
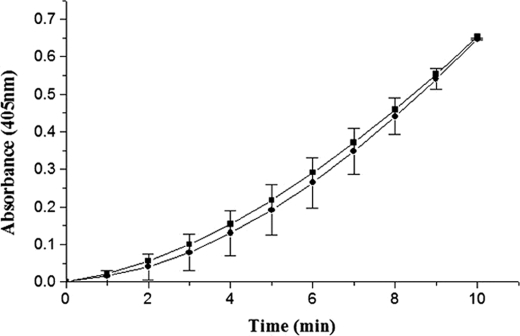
FIGURE 2.
Selective inhibition of substrate HPG activation ability of wtSK·HPN activator complex by 170 loop peptide mimotopes. Aliquots of preformed SK·HPN complex (5 nm) were added to HPG (0.05 μm) in assay buffer containing various concentrations of the 170 loop sequence peptides (0–500 μm). Residual percent activity in the presence of either linear, unconstrained 170 loop peptide (open circles), wild-type cyclic 170 loop peptide (solid squares), linear K180A peptide (solid circles), or the disulfide constrained cyclic K180A peptide (solid diamonds), was calculated from the rates of plasmin generated in the presence of peptide compared with that of activation reaction (taken as 100%) where no peptide was added (see “Experimental Procedures” for details). The control reactions where either plasmin inhibition (in the absence of SK) was tested in the presence of peptides are also depicted (upward triangles), or where possible inhibition of SK·HPN activator complex in the absence of substrate plasminogen (downward triangles) are also depicted.
Construction, Purification, and Functional Characterization of Alanine Substitution Mutants of the 170 Loop of the β-Domain of SK
The sequence of residues 170–181 of SK, which comprises the 170 loop in the β-domain of this tri-domain protein, consists of a stretch containing several positively and negatively charged residues (16, 25). To further explore the possible role of this loop in SK-mediated plasminogen activation, and also to identify its important residues if the loop were indeed important in HPG activation, alanine substitution mutations were introduced into the loop in the full-length SK expressed in E. coli BL21 (DE3) cells as inclusion bodies and purified to near-homogeneity (see “Experimental Procedures” for details). Far-UV CD spectra of the purified 170 loop mutants (data not shown) showed a close resemblance with that of the wtSK, which was suggestive of the absence of any major structural impairment caused by the loop mutations.
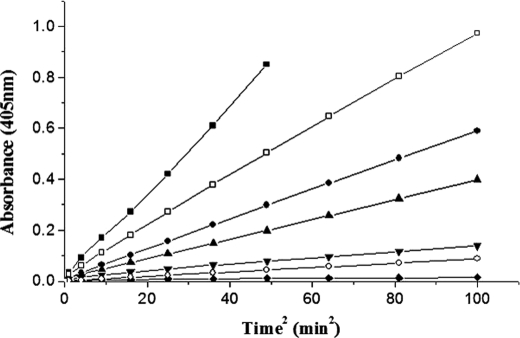
The mutants were then characterized for their ability to activate HPG by a single-stage continuous spectrophotometric assay that measures the overall HPG activator activity using catalytic amounts of SK and a large excess of HPG (28). The relative slopes of the progress curves so obtained (see Fig. 3, which shows the time course of plasmin generation by some of the alanine mutants compared with that of wtSK) clearly indicate that the alanine substitution mutants of 170 loop exhibited varied extents of reduction in their HPG activation capabilities. Of all the single-site substitution mutants tested, we observed that K180A exhibited the most compromised plasminogen activator activity, roughly of an order of magnitude less compared with wild-type SK. This was followed by P177A and R176A, carrying substitutions toward the central region of the loop. However, some mutants (D172A, D173A, and D174A) showed, at most, only a minor decrease in activity compared with wtSK.
FIGURE 3.
Time course of single stage HPG activation assays by wtSK and 170 loop mutants. SK/various 170 loop mutants of SK (0.5 nm each) were added individually to microtiter plate wells containing HPG and Chromozym®PL, and the activator activities were measured spectrophotometrically at 405 nm as detailed under “Experimental Procedures.” The graph shows wild-type SK (solid squares), R176A (solid circles), P177A (upward triangles), K180A (inverted triangles), K180G (open circles), K180D (solid diamonds), and D181A (open squares).
Because alanine substitution at position 180 in the full-length protein led to reduced plasminogen activation, it was of interest to perform direct inhibition assays with wtSK·HPN or mutant SK·HPN, as before, using the synthetic peptide approach. Using 170 loop wild-type peptide, both linear as well as cyclic, we checked for a possible inhibition of the mutant K180A·HPN enzyme complex, but none of these exhibited any significant inhibition at or near the concentrations seen to be inhibitory with wtSK·HPN (data not shown). This result suggests that the 170 loop sequence derived (wild-type) peptide, which likely mimics the specific conformation of the epitope present in the 170 loop of β-domain in streptokinase, loses its recognition (and, hence, capability for inhibition) once SK is mutated at the 180 position. To further explore the importance of this lysine to alanine substitution, linear as well as cyclic synthetic peptides, as before, carrying the alanine substitution at lysine 180 position were used to test for inhibition of wtSK·HPN enzyme complex in the HPG activation assays (Fig. 2; see also supplemental Fig. S1 for actual progress curves showing no inhibition by the mutated constrained peptide). From these results, it is apparent that the mutated constrained peptide had lost its ability for inhibition. Thus, overall, the results strongly support the conclusion that the 170 loop sequence represents a “hot spot” involved in substrate activation by SK·HPN.
Interaction of the 170 Loop Mutants of SK with Partner HPG/μPG
Because some of the single substitution mutants of the 170 loop exhibited significant reduction in their plasminogen activator capability, as monitored through single-stage assays, it was of interest to explore the underlying cause of this decline in activity, i.e. whether it was the amidolytic/zymogen activation step, which operates through Pathway I (schematically presented in Fig. 1) wherein 1:1 complexation of HPG with SK and formation of the SK·HPG* activator complex occurs, was affected, or a step subsequent to this, in Pathway II, namely the interaction of substrate HPG with the fully mature mutant SK·HPN complex. To address this issue, the ability of 170 loop mutants to generate the active site in HPG upon 1:1 complexation was examined through the well known reaction with the active site acylating agent, which is characterized by a rapid acylation of the active site in SK-HPG* marked by a burst of color liberation (24). This enables one to detect the formation of the activator complex even in the absence of proteolytic scission in the zymogen. Because amidolytic/esterolytic activation of HPG through Pathway I requires the presence of an intact N-terminal Ile (Ile1) in SK (19, 29, 30), it may be mentioned that the E. coli-derived, purified wtSK and the mutants had nearly 0.6 (14) mol of Ile per mole of protein as their N-terminal residue, which indicates a fairly high level of intracellular processing of the proteins by intrinsic E. coli fMet aminopeptidase (31). Subsequent methionine depletion of purified wtSK and K180A mutant under in vitro conditions was therefore carried out with MetAP (see “Experimental Procedures” for details). This resulted in further reduction of fMet to <0.1 mol/mol in both proteins. When these samples were reacted with NPGB, a nearly 2-fold jump in the extent of absorbance change for both wtSK and K180A mutant was observed (data not shown). The results presented in (Fig. 4) clearly show comparable acylation by the 170 loop mutant (K180A) and that of wtSK, which is in consonance with the results obtained through amidolytic activation of HPG, shown in supplemental Fig. S2. Closely similar results were obtained with two other mutants tested, namely SK K180G and SK K180D (data not shown).
FIGURE 4.
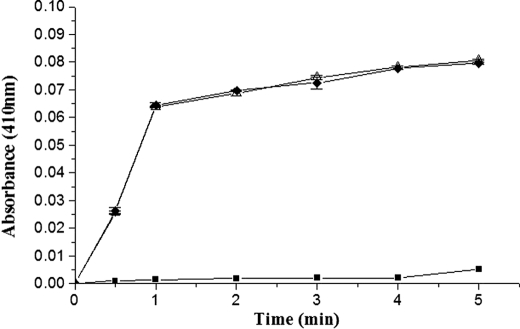
Active site titration of HPG upon complexing with wtSK/K180A mutant using the active site acylating agent, NPGB. The figure shows progress curves of NPGB hydrolysis as followed by p-nitrophenol liberation by MetAP-treated wtSK (open upward triangles), and MetAP-treated K180A (solid diamonds) complexes with HPG, and a control reaction (solid squares) where no activator was taken (see “Experimental Procedures” for details).
Likewise, the induction of a native-like complexation and opening of the active site in partner moiety through the use of the scissile peptide bond-mutated μPG (R561A), which does not transform to its plasminogen form, is also a test for Pathway I capability (19, 29). Past studies have shown that the R561A mutant of μPG can be activated by SK without being converted to μPN, and hence, can be utilized to rigorously test whether Pathway I capability is intact or compromised in a given SK mutant (19, 29). Accordingly, equimolar complexes of native SK or K180A with μPG R561A were tested (see “Experimental Procedures”) in a continuous spectrophotometric assay for amidolysis (Fig. 5). The results obtained clearly show the generation of nearly equivalent amidolytic activity upon addition of either K180A mutant or wtSK to R561A μPG mutant at 37 °C (Fig. 5).
FIGURE 5.
Time course of the amidolytic activity generation in R561A μPG by wtSK and K180A mutant. Equimolar SK·R561A μPG mutant and K180A·R561A μPG mutant complexes were made, and catalytic amounts were added to the assay buffer containing 1 mm chromogenic substrate Chromozym®PL, 50 mm Tris, pH 7.4, 100 mm NaCl, and amidolytic activity generated was represented as wild-type SK (solid squares) or K180A mutant (solid circles). Assays were performed at 37 °C with 10 nm SK and μPG complex as described under “Experimental Procedures.”
From these experiments, we can conclude that both wtSK as well as the 170 loop mutants have equivalent capability to generate esterolytic/amidolytic activity (NPGB hydrolysis) in partner HPG (see supplemental Fig. S2 for amidolytic activity generation in HPG), and therefore, the primary “functional lesion” due to 170 loop mutagenesis does not impair active site creation by SK in partner HPG moiety during the formation of a fully mature activator complex. This observation supports the conclusion that the reduced HPG activator capability observed in the mutant SK·HPN complexes arises from compromised enzyme-substrate interactions, rather than catalytic events related to 1:1 complexation of SK with HPG, and the maturation of the latter to the SK·HPN state.
Steady-state Kinetics of Plasminogen Activation by Equimolar Complexes of the 170 Loop Mutants with Plasmin Shows Selective Decline in Their Turnover Rates with No Concomitant Change in Enzyme-Substrate Affinity
To identify the underlying catalytic step that was affected in the various single substitution mutants of the 170 loop, steady-state kinetic studies were performed using a two-stage assay in which the Pathway II capability, i.e. the ability of the SK·HPN complex to act on substrate HPG is measured selectively (32, 33). Equimolar complexes of wtSK, or that of the individual 170 loop mutants, with HPN were first made, and their catalytic amounts were then employed as the enzyme complex for the activation of varying substrate HPG concentrations. The progress of each reaction was followed spectrophotometrically so as to obtain the steady-state kinetic parameters (28). The results are shown in Table 1. Interestingly, a nearly 10-fold drop in kcat was observed in the case of the K180A mutant without any significant change in the apparent affinity (Km) for substrate HPG (see supplemental Fig. S3 for the plots). It may be mentioned here that the plasminogen activator (specific) activity of the 170 loop mutant K180A remained compromised even when the mutant enzyme complex used to activate substrate HPG was at concentrations up to nearly 1000-fold higher than the concentrations usually employed in these assays (data not shown). If indeed the 1:1 affinity of the 170 loop mutants were compromised (and their low activity was a result of a non-optimal association of the mutant with plasmin), the specific activity obtained at say, the 100- and 1000-fold higher concentrations of the enzyme would have tended to become like that of wtSK. In contrast to K180A, the mutant P171A, and the ones in which the three consecutive aspartate residues were substituted with alanines, namely D172A, D173A, and D174A, exhibited near-native plasminogen activator activity. Once Pro177 was substituted with alanine, at the center of the loop, the resultant mutant also showed an 8- to 10-fold drop in activity. From these results, one can also infer that the proline side chain present in the central region of the loop likely plays a role in maintaining the functionality of this loop, because the deletion or alanine substitution of Pro177 perturbed the functionality of the loop. By contrast, proline at position 171 seemed to bear no functional relevance, because the specific activity of P171A mutant was essentially native-like. It is also noteworthy that, although the activation rates decreased to nearly one-tenth of that of wtSK by the lysine to alanine mutation at position 180, the mutation of the residue next to 180 (i.e. D181A) “allowed” the survival of nearly half the HPG activation capability seen with wtSK. This suggests that Lys180 has an important role to play in the functioning of the loop, along with Pro residue at position 177. Remarkably, the decrease in kcat seen with these mutants did not result in any significant alteration in apparent substrate affinity (to the extent that can be indicated by Km), which clearly suggests that the functional role of the 170 loop is related to generating substrate processivity in the SK·HPN enzyme complex rather than an overt role in macromolecular substrate binding/docking. These results clearly suggest that the central part, particularly residues in and around position 180, of the 170 loop is important for plasminogen activation.
TABLE 1.
Steady-state kinetic parameters for HPG activation by equimolar complexes of SK or the various 170 loop mutants and HPN
The kinetic parameters for substrate HPG activation were determined at 22 °C with the chromogenic substrate in 50 mm Tris-Cl buffer, pH 7.5, 100 mm NaCl as described under “Experimental Procedures.” The data represent the mean of three independent determinations.
| Activator species | Km | kcat | kcat/Km |
|---|---|---|---|
| μm | min−1 | min−1μm−1 | |
| wtSK | 0.40 ± 0.05 | 7.50 ± 0.3 | 19.00 |
| P171A | 0.40 ± 0.04 | 7.00 ± 0.5 | 17.50 |
| D172A | 0.50 ± 0.06 | 7.00 ± 0.5 | 14.0 |
| D173A | 0.50 ± 0.08 | 7.10 ± 0.4 | 14.2 |
| D174A | 0.50 ± 0.07 | 6.50 ± 0.3 | 13.0 |
| R176A | 0.60 ± 0.06 | 4.50 ± 0.4 | 7.5 |
| P177A | 0.45 ± 0.08 | 0.80 ± 0.03 | 1.7 |
| K180A | 0.50 ± 0.03 | 0.70 ± 0.01 | 1.40 |
| D181A | 0.55 ± 0.06 | 3.50 ± 0.34 | 6.3 |
| K180D | 0.50 ± 0.06 | 0.20 ± 0.06 | 0.4 |
| K180G | 0.60 ± 0.05 | 0.60 ± 0.04 | 1.0 |
| K180R | 0.50 ± 0.07 | 3.20 ± 0.50 | 6.4 |
| 177 pro del | 0.60 ± 0.05 | 0.80 ± 0.06 | 1.3 |
| R176A/K180A | 0.60 ± 0.08 | 0.70 ± 0.15 | 1.2 |
| R176A/D181A | 0.40 ± 0.07 | 4.00 ± 0.45 | 10 |
| P177A/K180A | 0.50 ± 0.07 | 0.70 ± 0.02 | 1.4 |
| K180A/D181A | 0.50 ± 0.06 | 0.80 ± 0.03 | 1.4 |
| R176A/K180A/D181A | 0.60 ± 0.08 | 0.40 ± 0.04 | 0.6 |
Various other double- and triple-alanine substitution mutants of the 170 loop were also constructed and examined for their capability to process substrate HPG (Table 1). Interestingly, from the results obtained, it became fairly evident that, although the presence of lysine at position 180 is important in the processing of substrate, other sites in the 170 loop are also contributive to this phenomenon. It can be seen that the mutation K180A, once in combination with other mutations within the 170 loop, such as in the triple substitution mutant, R176A/K180A/D181A, as well as the double substitution mutants, namely R176A/K180A and P177A/K180A, while essentially exhibiting the decline in catalytic turnover rates, still retained a substrate affinity (Km) that was essentially comparable to wild-type SK.
Probing the Importance of Lysine 180 in the 170 Loop of the β-Domain of Streptokinase
Because lysine to alanine mutation at position 180 exhibited a markedly reduced ability to activate substrate plasminogen by mutant SK·HPN activator complex, to substantiate our finding we examined whether a positively charged side chain at this position was mandatory for activation of HPG. Accordingly, we prepared different side-chain altered mutants at position 180 and examined their kinetics after complexation with HPN. Interestingly, from the results obtained (Table 1) it is evident that charge reversal mutation at lysine 180, to aspartate, significantly decreased the activation of HPG to levels ∼25-fold lower as compared with wtSK (Fig. 3). Glycine substitution at this position also resulted in a drop in specific activity that was roughly comparable to the alanine substitution mutant, but much better than in the case of the Lys to Asp mutation. Interestingly, when the residue was changed to arginine, a rejuvenation of the original activator activity was observed. Thus, the present study clearly highlights the importance of lysine 180 of the 170 loop of SK in catalytic processing of substrate plasminogen, which is likely due to the requirement of a positive charge at this position. Interestingly, the two amino acid residues identified to be relatively vital for 170 loop functioning, namely Pro177 and Lys180, are phylogenetically conserved across the various streptokinase allelic variant “classes” (Fig. 6) despite the generally diverse sequence heterogeneity in this region of streptokinase (34, 35). This suggests the evolutionary significance of these amino acid residues in the 170 loop that impart high catalytic turnover to SK·plasmin.
FIGURE 6.
Multiple alignment of primary sequence of β-domain depicting the 170 loop amino acid residues of streptokinases secreted from S. equisimilis and three representative allelic variant clusters of Streptococcus pyogenes, namely strains NS210, NS53, NS488, NS696, ALAB49, and NS13 (see Ref. 34). Note that residues (all given in single-letter code) highlighted in bold type are conserved in 170 loop in all the streptokinase polypeptides obtained from the different sources.
Selective Interaction of the 170 Loop of Streptokinase with the Catalytic Domain of Plasminogen
To obtain an idea of the broad locale in substrate plasminogen with which the 170 loop interacts, namely the catalytic domain or the kringle domains, a few key substitution mutants of 170 loop were studied for their ability to activate the isolated catalytic domain of plasminogen, i.e. μPG. We reasoned that if the mutants were affected in terms of kringle-mediated enzyme-substrate interactions only, their activity against μPG as substrate would be unchanged, i.e. it would be native-like. In contrast, if the mutants were compromised with respect to functioning of the exosites on SK that mediated enzyme-substrate interactions that were specifically targeted toward the catalytic domain per se, one would expect that their activity would be compromised similarly as observed with HPG as the substrate, in relative terms with respect to wtSK. It may be mentioned here that the activity of wtSK·HPN against μPG as the substrate is only ∼1–2% that of HPG, but once the full complement of the five kringle domains is present next to the catalytic domain (as in native HPG), the activator activity gets amplified to native-like levels (10). When the steady-state kinetic parameters for wtSK/loop mutants, after equimolar complexation with plasmin, were examined, the obtained results (Table 2) revealed a pattern of decreased catalytic turnover, compared with wtSK, which was closely similar in relative terms to that observed with full-length plasminogen as substrate. For example, in the case of the mutant K180A, which exhibits only one-tenth of the catalytic turnover rate compared with wtSK against native plasminogen as the substrate, nearly the same relative decline was seen in its catalytic rates for μPN generation from μPG, compared with the rates seen with wtSK. The same pattern of decline in μPG processing was observed in other alanine mutants of the 170 loop as well. All the alanine substitution mutants exhibited a Km of ∼1–2 μm, which is about the same as that exhibited by wtSK, but all of these mutants showed a pronounced decline in their ability to catalytically convert μPG to μPN, which was mostly similar to that seen when HPG was taken as the substrate. The proline to alanine substitution mutant, as well as the double mutant, P177A/K180A, showed a similar magnitude of decline in processing of μPG as that seen with HPG. Overall, these results suggest that the 170 loop of the β-domain of streptokinase encompasses a critical site of interaction between the SK·plasmin(ogen) complex and the catalytic domain of substrate plasminogen.
TABLE 2.
Steady-state kinetic parameters for μPG activation by equimolar complexes of SK/SK 170 loop mutants and HPN
The kinetic parameters for substrate μPG activation were determined at 22 °C with the chromogenic substrate in 50 mm Tris-Cl buffer, pH 7.5, 100 mm NaCl as described under “Experimental Procedures.” The data represent the mean of three independent determinations.
| Activator species | Km | kcat | kcat/Km |
|---|---|---|---|
| μm | min−1 | min−1μm−1 | |
| wtSK | 1.2 ± 0.1 | 0.16 ± 0.03 | 0.13 |
| K180A | 1.2 ± 0.4 | 0.01 ± 0.003 | 0.01 |
| K180G | 1.5 ± 0.1 | 0.01 ± 0.004 | 0.008 |
| K180R | 1.2 ± 0.1 | 0.06 ± 0.002 | 0.050 |
| K180D | 1.0 ± 0.2 | 0.01 ± 0.004 | 0.011 |
| P177A | 1.2 ± 0.3 | 0.02 ± 0.003 | 0.013 |
| P177A/K180A | 1.3 ± 0.2 | 0.01 ± 0.003 | 0.009 |
Real-time Surface Plasmon Resonance Studies for Ternary-mode Interactions of 170 Loop Mutants with Full-length and Truncated Substrates
The foregoing results for decreased kcat of the various 170 loop mutants for substrate HPG as well as μPG activation prompted us to explore whether the physicochemically measurable affinity of these substrates with preformed activator complexes between HPG and wtSK, on the one hand, and HPG with various 170 loop mutants, on the other, were affected or not using the real-time surface plasmon resonance technique. For this purpose, varying concentrations of substrate HPG were “docked” onto the preformed binary complex of immobilized HPG, and wtSK/various 170 loop mutants on the sensor surface, as described earlier (10, 14). The preformed SK·HPG complex has been shown to be relatively refractory to ϵ-aminocaproic acid, whereas the binding of the activator complex with the substrate HPG is susceptible to inhibition in the low millimolar range of concentration (36). The results obtained for the interaction of substrate HPG and μPG are given in Table 3. It is evident from the data that the equilibrium dissociation constants (KD) values for the ternary interaction of substrate HPG were closely similar to that observed earlier (14) in the case of ternary interaction of substrate HPG with native full-length SK (∼0.1 μm; see supplemental materials for binding sensorgrams). It is notable that, despite their lowered catalytic properties, the association and dissociation rate constants for ternary interactions of the 170 loop mutants were found to be closely similar to those observed in the case of ternary interaction of substrate HPG with wtSK.
TABLE 3.
Association (kon) and dissociation (koff) rate constants and apparent equilibrium dissociation (KD) constants for the interaction of substrate plasminogen and μPG with wtSK/SK loop mutants complexed with immobilized HPG by SPR
Kinetic constants for the interaction of substrate HPG and μPG with wtSK/loop mutants: HPG (binary complex) were determined by global fitting to a 1:1 binding model, using the BIAcore 3000 evaluation software as described under “Experimental Procedures.” A stable binary complex between wtSK/SK loop mutants and HPG immobilized onto the SA chip was first made, and the binding of varying concentrations of substrate HPG (0.05–1 μm), or substrate μPG (1–6 μm) was monitored (also see supplemental Figs. S2 and S3 for the sensorgrams).
| Ligand | Ligate HPG |
Ligate μPG |
||||
|---|---|---|---|---|---|---|
| kon | koff | KD | kon | koff | KD | |
| m−1s−1 × 105 | s−1 × 10−1 | m × 10−6 | m−1s−1 × 105 | s−1 × 10−1 | m × 10−6 | |
| wtSK·HPG | 12 ± 1.5 | 1.8 ± 0.2 | 0.15 ± 0.03 | 1.4 ± 0.5 | 1.5 ± 0.20 | 1.1 ± 0.3 |
| P177A·HPG | 8.0 ± 1.8 | 1.6 ± 0.2 | 0.20 ± 0.04 | 1.5 ± 0.3 | 1.7 ± 0.15 | 1.1 ± 0.2 |
| K180A·HPG | 11 ± 2.0 | 1.5 ± 0.3 | 0.14 ± 0.02 | 1.1 ± 0.3 | 1.4 ± 0.15 | 1.2 ± 0.2 |
| K180D·HPG | 12.5 ± 2.0 | 1.5 ± 0.4 | 0.12 ± 0.02 | 1.2 ± 0.2 | 1.2 ± 0.15 | 1.0 ± 0.2 |
When the docking of the isolated catalytic domain of plasminogen, i.e. μPG, was performed on the preformed binary complex of various 170 loop mutants with immobilized HPG, the results demonstrated a native-like equilibrium dissociation constant (KD) of the ternary interaction of substrate μPG with the three 170 loop mutants chosen for testing (Table 3). However, microplasminogen is known to be a poor substrate for the SK·plasmin(ogen) activator complex (14) with its reported KD for binding in the ternary mode with the preformed SK·HPG complex being in the range of 1–2 μm (14). Interestingly, the real-time affinity measurements using surface plasmon resonance analyses for the ternary interaction between μPG as the substrate and 170 loop mutant complexed with immobilized HPG show equilibrium dissociation constants (∼1.0 μm) that are nearly similar to that of wtSK. These results thus support, on a direct physicochemical level, the observations obtained from the steady-state kinetics studies that the 170 loop, although essential for catalytic processing of substrate plasminogen, does not seem to be contributive substantially toward enzyme-substrate affinity per se.
Even though the present study does not elucidate the exact molecular mechanism by which the 170 loop contributes to catalytic turnover, the observations are suggestive of the fact that the 170 loop interacts specifically with the catalytic domain of substrate. Surprisingly, this interaction does not seem to confer a high degree of enzyme-substrate affinity, because these mutants, after complexation with HPN/HPG, maintain a native-like docking potential for the macromolecular substrates but seem deficient in the post-docking phase of catalysis. These results complement a similar observation regarding the exclusivity of the substrate docking and turnover steps in this enzymatic system, wherein it was shown that the Asp41–His48 region of SK is involved in binding to substrate plasminogen independent of catalytic efficiency of its active binary complex (37).
The currently favored “exosite” model of SK action (4, 7, 9, 12, 14, 15, 36) envisions an “active valley” in the SK·HPN complex, with the trypsin-like plasmin active center located at its base. The involvement of exosite residues in proteinases of the blood coagulation pathway is well demonstrated in literature (38), an elegant example being that of the prothrombinase complex (39, 40), wherein substrate specificity, effector recognition, molecular activation, and regulation of thrombin (41, 42) are modulated by surface exosites unlike, say, a protease such as chymotrypsin, which acts primarily through the active site region (43). It is interesting to note that the role of the 170 loop-located exosite in SK action seems to be solely mediated through effects on the catalytic turnover of the SK·PN enzyme complex. In contrast, the exosite resident in the 250-loop seems to be operative also through direct substrate binding (14). It thus appears that SK possesses at least two types of exosites, namely one or more directed to substrate docking (characterized by “Km effects”), and “pure” kcat effects, as in the case of the 170 loop. The exact molecular mechanism whereby such exosites augment the catalysis by SK·HPN is not clear at this stage, but this may be through subtle changes in and around the active site of the SK-bound plasmin, which improve access to the substrate, or conversely, release of product, especially in the region around the scissile peptide bond in the macromolecular substrate. Thus, it is conceivable that the HPN active center remains essentially as “non-committal” with respect to its primary substrate specificity as is free HPN, but the overall SK·plasmin complex acquires a selective substrate-processing capacity through the stereochemically guided docking of the macromolecular substrate into this valley. Once this is accomplished, it is logical to assume that facile proteolytic cleavage could now occur at the substrate's scissile peptide bond, triggering the product expulsion step of catalysis. The exact identity of all exosites participating in this interaction, which also involves long range enzyme-substrate interactions as well (10), is not known currently. Nevertheless, this model is consistent with a mechanistic scenario wherein the 170 loop or, indeed, any other such surface-located, substrate-interacting site(s) on SK, such as the 250-loop or the 88–97 loop (12, 14) act as flexible pivots, or hinges, that facilitate the rapid release of the product from the enzyme at the end of each catalytic cycle even if such an interaction may, or may not (as in case of the 170 loop) necessarily be contributive in terms of enhancing enzyme-substrate affinity. Indeed, one such site, which does contribute toward enzyme-substrate affinity, has already been identified to reside in the fifth kringle of HPG (14, 15). However, the interacting site for the 170 loop in substrate HPG now needs to be identified to closely examine the dynamics of their interaction. Efforts along these lines, using fluorescence resonance energy transfer and cross-linking studies, are currently underway in our laboratory. Besides SK, the role of surface-located loops in plasminogen activation has been strongly indicated in other bacterial and physiological activators as well, such as staphylokinase, tissue plasminogen activator, and urokinase plasminogen activator (44–46), but the exact manner whereby such loops modulate enzymatic specificity and/or catalysis remains to be satisfactorily elucidated. Such an understanding, undoubtedly, will be helpful in the redesign of existing plasminogen activators, and perhaps, also in the de novo design of proteases with new substrate specificities. In our current working model of SK action, it is the co-operative action of the various exosites that leads to the generation of high rates of plasminogen activation by SK, which is known to be highest among all other physiological and bacterial PG activators (47). Our preliminary observations3 with simultaneous multisite mutations in the various exosites of SK identified so far, on the one hand, and sites in the macromolecular substrate that are appreciably distant from the scissile peptide bond, on the other, reveal highly synergistic effects on the catalytic rates for HPG activation. These observations are intriguingly indicative of a catalytic mechanism that is centered on the existence of a “distributed” network of exosites in the SK·HPN enzymatic complex.
Supplementary Material
Acknowledgments
We thank Paramjit Kaur and Deepak Bhatt for expert technical support, and the Blood Bank, Government Medical College, Chandigarh for human plasma.
This work was supported by intra-mural funds from The Institute of Microbial Technology, India.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5 and Table SI.
R. Aneja, S. Yadav, K. Joshi, S. Kumar, and G. Sahni, unpublished observations.
- SK
- streptokinase
- μPG
- microplasminogen
- μPN
- microplasmin
- HPG
- human plasminogen
- HPN
- human plasmin
- koff
- rate of dissociation
- kon
- rate of association
- MetAP
- methionine aminopeptidase
- NPGB
- p-nitrophenyl p-guanidinobenzoate
- wtSK
- wild-type streptokinase.
REFERENCES
- 1.Lijnen H. R., Stassen J. M., Vanlinthout I., Fukao H., Okada K., Matsuo O., Collen D. (1991) Thromb. Haemost. 66, 468–473 [PubMed] [Google Scholar]
- 2.McClintock D. K., Bell P. H. (1971) Biochem. Biophys. Res. Commun. 43, 694–702 [DOI] [PubMed] [Google Scholar]
- 3.Bajaj A. P., Castellino F. J. (1977) J. Biol. Chem. 252, 492–498 [PubMed] [Google Scholar]
- 4.Boxrud P. D., Verhamme I. M., Bock P. E. (2004) J. Biol. Chem. 279, 36633–36641 [DOI] [PubMed] [Google Scholar]
- 5.Boxrud P. D., Bock P. E. (2004) J. Biol. Chem. 279, 36642–36649 [DOI] [PubMed] [Google Scholar]
- 6.Markus G., Werkheiser W. C. (1964) J. Biol. Chem. 239, 2637–2643 [PubMed] [Google Scholar]
- 7.Boxrud P. D., Fay W. P., Bock P. E. (2000) J. Biol. Chem. 275, 14579–14589 [DOI] [PubMed] [Google Scholar]
- 8.Reddy K. N., Markus G. (1972) J. Biol. Chem. 247, 1683–1691 [PubMed] [Google Scholar]
- 9.Wang X., Lin X., Loy J. A., Tang J., Zhang X. C. (1998) Science 281, 1662–1665 [DOI] [PubMed] [Google Scholar]
- 10.Sundram V., Nanda J. S., Rajagopal K., Dhar J., Chaudhary A., Sahni G. (2003) J. Biol. Chem. 278, 30569–30577 [DOI] [PubMed] [Google Scholar]
- 11.Chaudhary A., Vasudha S., Rajagopal K., Komath S. S., Garg N., Yadav M., Mande S. C., Sahni G. (1999) Protein Sci. 8, 2791–2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yadav S., Datt M., Singh B., Sahni G. (2008) Biochim. Biophys. Acta. 1784, 1310–1318 [DOI] [PubMed] [Google Scholar]
- 13.Reed G. L., Lin L. F., Parhami-Seren B., Kussie P. (1995) Biochemistry 34, 10266–10271 [DOI] [PubMed] [Google Scholar]
- 14.Dhar J., Pande A. H., Sundram V., Nanda J. S., Mande S. C., Sahni G. (2002) J. Biol. Chem. 277, 13257–13267 [DOI] [PubMed] [Google Scholar]
- 15.Tharp A. C., Laha M., Panizzi P., Thompson M. W., Fuentes-Prior P., Bock P. E. (2009) J. Biol. Chem. 284, 19511–19521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X., Tang J., Hunter B., Zhang X. C. (1999) FEBS Lett. 459, 85–89 [DOI] [PubMed] [Google Scholar]
- 17.Deutsch D. G., Mertz E. T. (1970) Science 170, 1095–1096 [DOI] [PubMed] [Google Scholar]
- 18.Shi G. Y., Wu H. L. (1988) J. Biol. Chem. 263, 17071–17075 [PubMed] [Google Scholar]
- 19.Wang S., Reed G. L., Hedstrom L. (1999) Biochemistry 38, 5232–5240 [DOI] [PubMed] [Google Scholar]
- 20.Radek J. T., Davidson D. J., Castellino F. J. (1993) Methods Enzymol. 223, 145–155 [DOI] [PubMed] [Google Scholar]
- 21.Wohl R. C., Summaria L., Robbins K. C. (1980) J. Biol. Chem. 255, 2005–2013 [PubMed] [Google Scholar]
- 22.Wakeham N., Terzyan S., Zhai P., Loy J. A., Tang J., Zhang X. C. (2002) Protein Eng. 15, 753–761 [DOI] [PubMed] [Google Scholar]
- 23.Chase T., Jr., Shaw E. (1969) Biochemistry 8, 2212–2224 [DOI] [PubMed] [Google Scholar]
- 24.Wohl R. C. (1984) Biochemistry 23, 3799–3804 [DOI] [PubMed] [Google Scholar]
- 25.Jackson K. W., Tang J. (1982) Biochemistry 21, 6620–6625 [DOI] [PubMed] [Google Scholar]
- 26.Nihalani D., Raghava G. P., Sahni G. (1997) Protein Sci. 6, 1284–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nihalani D., Kumar R., Rajagopal K., Sahni G. (1998) Protein Sci. 7, 637–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Renzo E. C., Siiteri P. K., Hutchings B. L., Bell P. H. (1967) J. Biol. Chem. 242, 533–542 [PubMed] [Google Scholar]
- 29.Wang S., Reed G. L., Hedstrom L. (2000) Eur. J. Biochem. 267, 3994–4001 [DOI] [PubMed] [Google Scholar]
- 30.Boxrud P. D., Verhamme I. M., Fay W. P., Bock P. E. (2001) J. Biol. Chem. 276, 26084–26089 [DOI] [PubMed] [Google Scholar]
- 31.Hirel P. H., Schmitter M. J., Dessen P., Fayat G., Blanquet S. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 8247–8251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Summaria L., Wohl R. C., Boreisha I. G., Robbins K. C. (1982) Biochemistry 21, 2056–2059 [DOI] [PubMed] [Google Scholar]
- 33.Gonzalez-Gronow M., Siefring G. E., Jr., Castellino F. J. (1978) J. Biol. Chem. 253, 1090–1094 [PubMed] [Google Scholar]
- 34.Lizano S., Johnston K. H. (2005) Infect Immun. 73, 4451–4453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McArthur J. D., McKay F. C., Ramachandran V., Shyam P., Cork A. J., Sanderson-Smith M. L., Cole J. N., Ringdahl U., Sjöbring U., Ranson M., Walker M. J. (2008) FASEB J. 22, 3146–3153 [DOI] [PubMed] [Google Scholar]
- 36.Lin L. F., Houng A., Reed G. L. (2000) Biochemistry 39, 4740–4745 [DOI] [PubMed] [Google Scholar]
- 37.Kim D. M., Lee S. J., Kim I. C., Kim S. T., Byun S. M. (2000) Thromb. Res. 99, 93–98 [DOI] [PubMed] [Google Scholar]
- 38.Bock P. E., Panizzi P., Verhamme I. M. (2007) J. Thromb. Haemost. 5, Suppl. 1, 81– 94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boskovic D. S., Krishnaswamy S. (2000) J. Biol. Chem. 275, 38561–38570 [DOI] [PubMed] [Google Scholar]
- 40.Krishnaswamy S. (2005) J. Thromb. Haemost. 3, 54–67 [DOI] [PubMed] [Google Scholar]
- 41.Huntington J. A. (2005) J. Thromb. Haemost. 3, 1861–1872 [DOI] [PubMed] [Google Scholar]
- 42.Bode W. (2006) Blood Cells Mol. Dis. 36, 122–130 [DOI] [PubMed] [Google Scholar]
- 43.Furie B., Bing D. H., Feldmann R. J., Robison D. J., Burnier J. P., Furie B. C. (1982) J. Biol. Chem. 257, 3875–3882 [PubMed] [Google Scholar]
- 44.Rajamohan G., Dahiya M., Mande S. C., Dikshit K. L. (2002) Biochem. J. 365, 379–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tachias K., Madison E. L. (1996) J. Biol. Chem. 271, 28749–28752 [DOI] [PubMed] [Google Scholar]
- 46.Petersen H. H., Hansen M., Schousboe S. L., Andreasen P. A. (2001) Eur. J. Biochem. 268, 4430–4439 [DOI] [PubMed] [Google Scholar]
- 47.Lee P. P., Wohl R. C., Boreisha I. G., Robbins K. C. (1988) Biochemistry 27, 7506–7513 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.