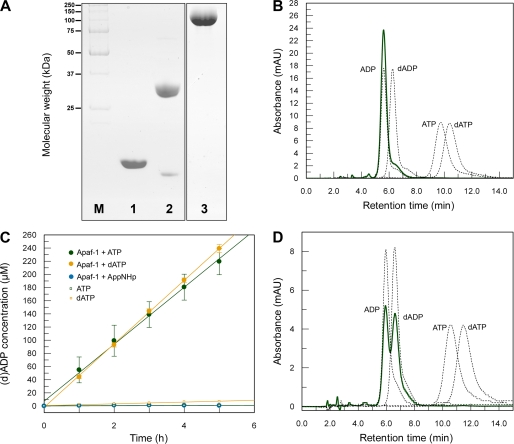
FIGURE 1.
Nucleotide status and hydrolytic properties of Apaf-1. A, Coomassie-stained SDS-PAGE showing the purity of the proteins used in the assays. Lane M, molecular weight marker; lane 1, horse cytochrome c; lane 2, caspase-9; lane 3, Apaf-1. B, Apaf-1 purified from Sf21 insect cells contains ADP. Absorption curves corresponding to reference nucleotides are shown as black dotted lines; the absorption curve of the nucleotide isolated from purified Apaf-1 is shown as green solid line. C, Apaf-1 hydrolyzes ATP and dATP at a low rate. 10 μm Apaf-1 was incubated in buffer A at 30 °C with 1 mm ATP and dATP, respectively. Samples taken at different time points were analyzed on an HPLC system to detect the formed nucleoside diphosphate. By comparison with reference samples, the respective peak area (supplemental Fig. S1) was converted into concentration (green or yellow dots). As controls, the same was done with samples containing nucleotide but no Apaf-1 (green or yellow open squares) or Apaf-1 together with the nonhydrolyzable ATP analog AppNHp (blue dots). Mean averages of triplicate measurements are plotted against time. The resulting linear fits represent an ATPase activity of 4.33 ± 0.13 ATP molecules per molecule of Apaf-1 per h and a dATPase activity of 4.83 ± 0.04 dATP molecules per molecule of Apaf-1 per h. D, Apaf-1 is able to exchange ADP for dATP in the absence of cytochrome c. 500 μg of Apaf-1 were incubated with 1 mm dATP at 20 °C for 3 h. After separation of the protein from excess nucleotide by gel filtration, the peak fractions were concentrated and analyzed by HPLC. Absorption curves corresponding to reference nucleotides are shown as black dotted lines, and the absorption curve of the nucleotide isolated from Apaf-1 is shown as green solid line. mAU, milliabsorbance units.