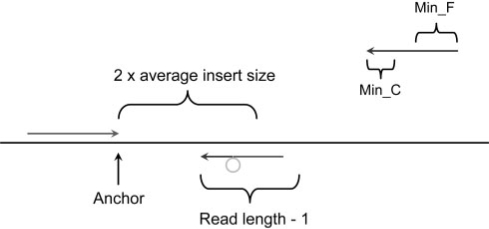
Fig. 2.
Detecting short insertion events. The procedure to split the unmapped read into three parts at appropriate position and mapped the terminal two separately to the reference genome. The location and direction of the mapped read (green) define the local region to split the unmapped read. The 3′ end of the mapped read is defined as anchor point. Then pattern growth is used to search for minimum and maximum unique substrings from the 3′ end of unmapped reads within the range of two times of insert size starting from the anchor point. Using pattern growth again to search for minimum and maximum unique substrings from the 5′ of unmapped read within the range of read length – 1, starting from the already mapped 3′ end of the unmapped read. The computed minimum and maximum substrings from both 3′ and 5′ are examined to see whether they are adjacent to each other. The middle fragment is the inserted fragment. All possible solutions are stored in a database for sorting according to the break point coordinates. An insertion event is reported if at least two reads support it.