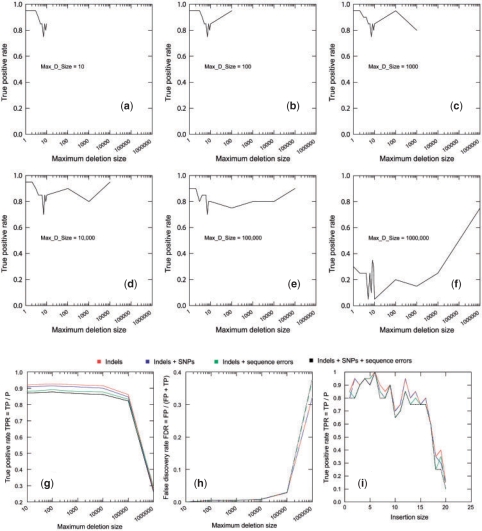
Fig. 4.
Simulation of paired-end reads from human chromosome X. (a–f) True positive rates per each deletion size are displayed in the presence of sequencing errors and SNPs when Max_D_Size is increased from 10 to 1 000 000 bp. The impact of sequencing errors and/or SNPs on the overall true positive rates (g) and false discovery rates (h) when Max_D_Size is set to different values from 10 to 1 000 000. (i) The impact of sequencing errors and/or SNPs on the true positive rates for detecting medium sized insertions from size 1 to –20 bp.