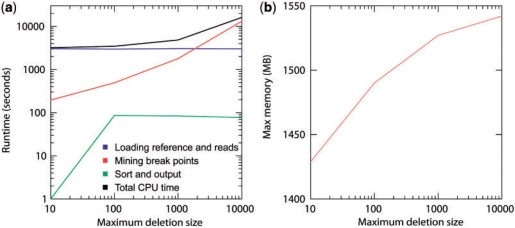
Fig. 6.
Runtime and memory consumption for Pindel applied to the NA18507 data on a single CPU for mining indels with different Max_D_Size (10 bp, 100 bp, 1 kb and 10 kb). (a) The user runtime for Pindel is divided into three categories: (i) loading the reference genome and the reads into memory. (ii) Break unmapped reads and map them separately using pattern growth. (iii) Sort break points according to coordinates and write the results on hard disk. (b) Maximum memory consumption for Pindel to process the NA18507 data with different Max_D_Size (10 bp, 100 bp, 1 kb and 10 kb).