Abstract
Summary: The increasing availability of large network datasets along with the progresses in experimental high-throughput technologies have prompted the need for tools allowing easy integration of experimental data with data derived form network computational analysis. In order to enrich experimental data with network topological parameters, we have developed the Cytoscape plug-in CentiScaPe. The plug-in computes several network centrality parameters and allows the user to analyze existing relationships between experimental data provided by the users and node centrality values computed by the plug-in. CentiScaPe allows identifying network nodes that are relevant from both experimental and topological viewpoints. CentiScaPe also provides a Boolean logic-based tool that allows easy characterization of nodes whose topological relevance depends on more than one centrality. Finally, different graphic outputs and the included description of biological significance for each computed centrality facilitate the analysis by the end users not expert in graph theory, thus allowing easy node categorization and experimental prioritization.
Availability: CentiScaPe can be downloaded via the Cytoscape web site: http://chianti.ucsd.edu/cyto_web/plugins/index.php. Tutorial, centrality descriptions and example data are available at: http://profs.sci.univr.it/∼scardoni/centiscape/centiscapepage.php
Contact: giovanni.scardoni@gmail.com
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
The vast amount of available experimental data generating annotated gene or protein complex networks has increased the quest for network analysis. Biological networks are usually represented as graphs, where the nodes are biological entities (such as cells, genes, proteins or metabolites) and the edges are functional and/or physical interactions between them. Visualization and analysis tools are needed to understand individual node functions masked by the overall network complexity. Several techniques suitable to network structural analysis exist, such as the analysis of the global network structure (Albert and Barabasi, 2002), network motifs (Milo et al., 2002), network clustering (Holme et al., 2003) and network centralities (Wutchy and Stadler, 2003). Particularly, centralities are node parameters that can identify nodes having a relevant position in the overall network architecture. Cytoscape (Cline et al., 2007; Shannon et al., 2003) is an excellent visualization and analysis tool with the analysis features greatly enhanced by plug-ins. Plug-ins such as NetworkAnalyzer (Assenov et al., 2008) computes some node centralities but does not allow direct integration with experimental data. Applications such as VisANT (Hu et al., 2005) and Centibin (Junker et al., 2006) calculate centralities, although they either calculate fewer centralities or are not suitable to integration with experimental data (see Supplementary Table S1 for a comparative evaluation). CentiScaPe is the only Cytoscape plug-in that computes several centralities at once. In CentiScaPe, computed centralities can be easily correlated between each other or with biological parameters derived from the experiments in order to identify the most significant nodes according to both topological and biological properties. Functional to this capability is the scatter plot by value options, which allows easy correlating node centrality values to experimental data defined by the user.
2 SYSTEM OVERVIEW
CentiScaPe computes several network centralities only for undirected networks. Computed parameters are: Average Distance, Diameter, Degree, Stress, Betweenness, Radiality, Closeness, Centroid Value and Eccentricity. Plug-in help and online files are provided with definition, description, biological significance and computational complexity for each centrality (Supplementary Tables S2 and S3, CentralitiesTutorial). Min, max and mean values are given for each computed centrality. Multiple networks analysis is also supported. Centrality values appear in the Cytoscape attributes browser, so they can be saved and loaded as normal attributes, thus allowing their visualization with the Cytoscape mapping core features. Once computation is completed, the actual analysis begins, using the graphical interface of CentiScaPe. CentiScaPe uses the free Java libraries JFreeChart (http://jfree.org/jfreechart/) to display the results as graphical outputs. The first step of the analysis is the Boolean logic-based result panel of CentiScaPe. It is possible, by using the provided sliders in the Results Panel of Cytoscape, to highlight the nodes having centralities values that are higher, minor or equal to a threshold value defined by the user (the mean value is used by default). If necessary, one or more centralities can be deactivated. The user can select the more/equal option for some centralities, the less/equal option for others and can join them with AND-OR operators. This feature can immediately answer to questions as: ‘Which are the nodes having high Betweenness and Stress but low Eccentricity?’ Notably, the threshold can also be modified by hand to gain in resolution. Once the nodes have been selected according to their node-specific values, the corresponding subgraph can be extracted and displayed using normal Cytoscape core features. Two kind of graphical outputs are supported: plot by centrality and plot by node, both allowing analysis that are not possible with other centralities tools. The user can correlate centralities between them or with experimental data, such as, for example, gene expression level or protein phosphorylation level (plot by centralities), and can analyze all centralities values node by node (plot by node) (Fig. 1). Graphics can be saved to a jpeg file.
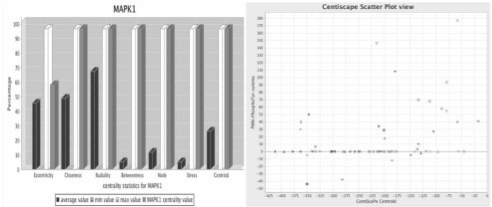
Fig. 1.
Network analysis of human kino-phosphatome. (A) The protein kinase MAPK1 shows high centralities values for most of the computed centralities suggesting its central role in the network structure and function. For each centrality the specific node value (gray), the mean value (black), the min value (light gray) and the max value (white) is shown. (B) The correlations between centroid value and intensity of protein phosphorylation in Tyrosine. Proteins with high centroid value and high level of phosphorylation are easily identified in the top/right quadrant of the graph. Pointing the mouse over the geometrical shapes in the plot shows the corresponding node ID and attribute values (see Section 3 and Supplementary Materials).
The plot by centrality visualization is an easy and convenient way to discriminate nodes and/or group of nodes that are most relevant according to a combination of two selected parameters. It shows correlation between centralities and/or other quantitative node attributes, such as experimental data from genomic and/or proteomic analysis. The result of the plot by centrality option is a chart where each individual node, represented by a geometrical shape, is mapped to a Cartesian axis. In the horizontal and vertical axis, the values of the selected attributes are reported. Most of the relevant nodes are easily identified in the top-right quadrant of the chart. Figure 1 (Supplementary Fig. S1) shows a plot of centroid values over intensity of protein tyrosine phosphorylation in the human kino-phosphatome network derived from the analysis of human primary polymorphonuclear neutrophils (PMNs) stimulated with the chemoattractant IL-8 (Section 3). The proteins having high values for both parameters likely play a crucial regulatory role in the network. The user can plot in five different ways: centrality versus centrality, centrality versus experimental data, experimental data versus experimental data, a centrality versus itself and an experimental data versus itself. Notably, a specific way to use the plot function is to visualize the scatter plot of two experimental data attributes. This is an extra function of the plug-in and can be used in the same way of the centrality/centrality option and centrality/experimental attribute option. If the plot by centrality option is used selecting the same centrality (or the same experimental attribute) for both the horizontal and the vertical axis, result is an easy discrimination of nodes having low values from nodes having high values of the selected parameter. Thus, the main use of the ‘plot by centrality’ feature is to identify group of nodes clustered according to combination of specific topological and/or experimental properties, in order to extract sub-networks to be further analyzed. The combination of topological properties with experimental data is useful to allow more meaningful predictions of sub-network function to be experimentally validated.
The plot by node option, another unique feature of CentiScaPe, shows for every single node the value of all calculated centralities represented as a bar graph. The mean, max and min values are represented with different colors. To facilitate the visualization, all the values in the graph are normalized and the real values appear when pointing the mouse over a bar. Figure 1 (Supplementary Fig. S2) shows, as an example, the values for the MAPK1 calculated from the global human kino-phosphatome.
3 A REAL WORLD EXAMPLE: CENTRALITIES IN THE HUMAN KINO-PHOSPHATOME
We tested CentiScaPe on the human kino-phosphatome. A global human protein interactome dataset, including 11 120 nodes and 84 776 unique undirected interactions (IDs=HGNC), was complied from public databases (HPRD, BIND, DIP, IntAct, MINT, BioGRID; Supplementary file GLOBAL-HGNC.sif). A subset of this network was extracted consisting of only known interactions between human protein kinases and phosphatases. The resulting sub-network, a kino-phosphatome network, consisted of 549 nodes and 3844 unique interactions (Supplementary Table S4 and Kino-Phosphatome.sif), with 406 kinases and 143 phosphatases. The kino-phosphatome network did not contain isolated nodes. We used CentiScaPe to calculate centrality parameters. A first general overview of the global topological properties of the kino-phosphatome network comes from the min, max and average values of all computed centralities along with the diameter and the average distance of the network (Supplementary Table S5). For instance, an average degree =13.5 with an average distance of 3 may suggest a highly connected network, in which proteins are strongly functionally interconnected. Computation of network centralities allowed a first ranking of human kinases and phosphatases according to their central role in the network (Supplementary Table S6, reporting node-by-node values of different centralities). To facilitate the identification of nodes with the highest scores we applied the ‘plot by centrality’ feature of CentiScaPe. Plotting degree over degree (Supplementary Fig. S3) shows that the distribution is not uniform, with the majority of nodes having a similar low degree and very few having very high degree. This is consistent with the known scale-free architecture of biological networks (Jeong et al., 2000). The scale-free topology of the kino-phosphatome network was also confirmed with Network Analyzer (Assenov et al., 2008). A total of 186 nodes (164 kinases and 22 phosphatases) displayed a degree over the average. The top-10 degree values (64–102) were all kinases, with MAPK1 showing the highest degree (102). Notably, MAPK1 displayed the highest score for all computed centralities (Fig. 1), suggesting its central regulatory role in the kino-phosphatome. In contrast, PTPN1 had the highest degree, 46, between all phosphatases (top 31 among all nodes) and had a rather high score also for other centralities (Supplementary Fig. S4). Thus, degree analysis suggests that MAPK1 and PTPN1 are the most central kinase and phosphatase, respectively.
To further support this suggestion we analyzed also the centroid. Plotting centroid over centroid provided a linear distribution and as for the degree, also here the distribution was not uniform (Supplementary Figs S5 and S3). Average centroid was -393. A total of 242 nodes (206 kinases and 36 phosphatases) displayed a centroid over the average. The top-10 centroid values (-79 to 18) were all kinases, with MAPK1 showing the highest centroid value (18). PTPN1 had the highest centroid value, -154, between all phosphatases (top 22 among all nodes). Thus, as for the degree, also the centroid value analysis suggests a scale-free distribution, with MAPK1 and PTPN1 being the most central kinase and phosphatase, respectively. This conclusion was also easily evidenced by plotting the degree over the centroid (Supplementary Fig. S6). From the analysis, a non-linear distribution of nodes is evident, with few dispersed nodes occupying the top-right quadrant of the plot (i.e. high degree and high centroid): these nodes can potentially represent particularly important regulatory kinases and phosphatases.
This kind of analysis can be iterated by evaluating all other centralities. To extract the most relevant nodes, we used CentiScaPe to select all nodes having all centrality values over the average. Upon filtering, we obtained a kino-phosphatome sub-network consisting of 97 nodes (82 kinases and 15 phosphatases) and 962 interactions (Supplementary Fig. S7, Table S7 and K-P sub-network.sif). This sub-network possibly represents a group of highly interacting kinases and phosphatases displaying a critical role in the regulation of protein phosphorylation in human cells. Further analysis with CentiScaPe or other analysis tools, such as MCODE (Bader and Hogue, 2003) or network analyzer, performing a Gene Ontology (Ashburner et al., 2000) database search, or adding functional annotation data, may allow a deeper functional exploration of the sub-network.
The regulatory role of proteins in the kino-phosphatome network may be also experimentally tested in a context-selective manner. Indeed, the centrality analysis by CentiScaPe can be even more significant by superimposing experimental data. To test this possibility, we focused the analysis on human PMNs (Supplementary file Phosphorylation-Experiment for description). Human neutrophils were stimulated under stirring at 37○C for 1 min with the classical chemoattractant fMLP (100 nM) and protein phosphorylation was evaluated by using the Kinexus protein array service (www.kinexus.ca) (phosphorylation data are available in Supplementary Material: PMN-PhosphoSer.NA, PMN-PhosphoTyr.NA, PMN-PhosphoThr.NA). Experimental data were loaded as node attributes in Cytoscape and the computed centrality values were plotted over values of protein phosphorylation. Here, every node is represented with two coordinates consisting of computed centrality and of experimental data regarding protein phosphorylation induced in PMNs by fMLP. Plot of centroid values over intensity of protein phosphorylation in threonine or tyrosine residues induced by fMLP triggering in human PMNs have been analyzed. The plots allow immediately evidencing that proteins phosphorylated in threonine (Supplementary Fig. S8) or in tyrosine (Fig. 1 and Supplementary Fig. S1) have different topological position in the network with proteins phosphorylated in tyrosine showing a higher centrality values. This could suggest that tyrosine phosphorylation induced in PMNs by chemoattractants involves signaling proteins regulating clusters of proteins, as the centroid value may suggest. Further hypotheses can be formulated by expanding the analysis to other centralities and by expanding the phopshorylation data. From this type of plotting it is possible to further identify relevant nodes not only according to topological position but also to experimental outputs. Thus, groups of nodes whose regulatory relevance is suggested by centrality analysis are further characterized by the corresponding data of biological activity. In this context, the topological analysis and experimental data do confirm each other's regulatory relevance and may suggest further, more focused, experimental verifications. Combination of CentiScaPe with other bioinformatics tools may help to analyze high-throughput genomic and/or proteomic experimental data and may facilitate the decision process.
4 CONCLUSIONS
CentiScaPe is a versatile and user-friendly bioinformatic tool to integrate centrality-based network analysis with experimental data. CentiScaPe is completely integrated into Cytoscape and the possibility of treating centralities as normal attributes permits to enrich the analysis with the Cytoscape core features and with other Cytoscape plug-ins. The analysis obtained with the Boolean-based result panel, the ‘plot by node’ and the ‘plot by centrality’ options give meaningful results not accessible to other tools and allow easy categorization of nodes in large complex networks derived from experimental data.
Funding: Fondazione Cariverona (to CBMC); Fondazione Cariverona (to C.L.); Associazione Italiana per la Ricerca sul Cancro (to C.L.).
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Albert R, Barabasi AL. Statistical mechanics of Complex Network. Rev. Mod. Phys. 2002;74:47–97. [Google Scholar]
- Ashburner M, et al. Gene Ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assenov Y, et al. Computing topological parameters of biological networks. Bioinformatics. 2008;24:282–284. doi: 10.1093/bioinformatics/btm554. [DOI] [PubMed] [Google Scholar]
- Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline MS, et al. Integration of biological networks and gene expression data using Cytoscape. Nat. Protocols. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holme P, et al. Subnetwork hierarchies of biochemical pathways. Bioinformatics. 2003;19:532–538. doi: 10.1093/bioinformatics/btg033. [DOI] [PubMed] [Google Scholar]
- Hu Z, et al. VisANT: data integrating visual framework for biological networks and modules. Nucleic Acid Res. 2005;33:W352–W357. doi: 10.1093/nar/gki431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H, et al. The large-scale organization of metabolic networks. Nature. 2000;407:651–655. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- Junker BH, et al. Exploration of biological network centralities with CentiBiN. BMC Bioinformatics. 2006;7:219. doi: 10.1186/1471-2105-7-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milo R, et al. Network Motifs: simple building blocks of complex networks. Science. 2002;298:824. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutchy S, Stadler PF. Centers of complex networks. J. Theor. Biol. 2003;223:45–53. doi: 10.1016/s0022-5193(03)00071-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.