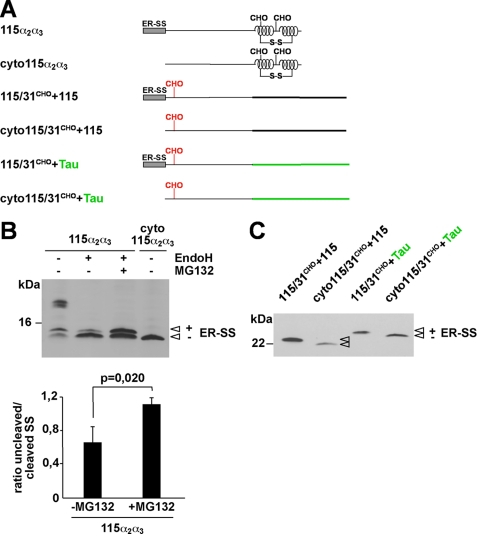
FIGURE 4.
Polypeptides subjected to proteasomal degradation contain an uncleaved signal sequence. A, schematic presentation of the mutants analyzed. Two versions of the mutants 115α2α3, 115/31CHO+115, and 115/31CHO+Tau were generated, one version with the original ER-SS and one lacking the ER-SS (cyto forms). B, N2a cells were transiently transfected and incubated in the presence or absence of MG132 (30 μm; 3 h). In addition, cell lysates were either treated with EndoH (+ EndoH) or left untreated (− EndoH) prior to Western blotting using the mAb 3F4. Unglycosylated PrP with (+ ER-SS) and without (− ER-SS) the ER signal peptide are marked. Lower panel, quantification of at least three independent experiments. Plotted is the ratio of the amount of 115α2α3 with an uncleaved versus cleaved SS in EndoH-treated samples with or without proteasomal inhibition (± MG132) (mean ± S.E.). The p value was determined by Student's t test. C, N2a cells transfected with the mutants depicted were lysed, and proteins analyzed by Western blot using the mAb 3F4. The protein fraction with (+ ER-SS) and without (− ER-SS) the ER signal peptide is marked.