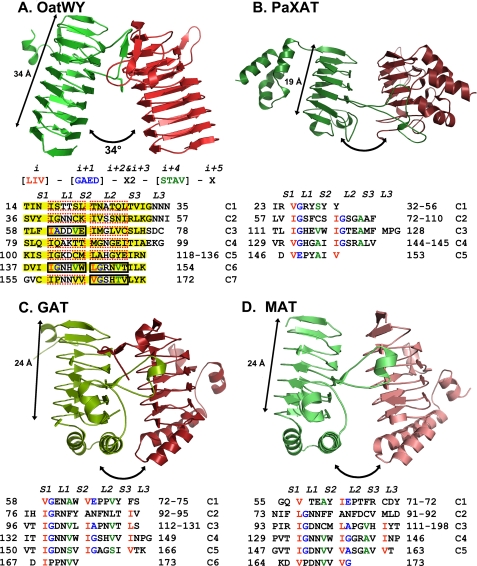
FIGURE 4.
Structural comparison of OatWY with other bacterial acetyltransferases containing LβH motifs (A, polysialic acid O-acetyltransferase OatWY; B, xenobiotic acetyltransferase PaXAT (PDB access code 2XAT); C, GAT (PDB access code 1KRU); D, MAT (PDB access code 1OCX)). Only two monomeric subunits of the trimer are represented for clarity (in green and red). Angles between the 3-fold axis and LβH domains are illustrated with two-headed arrows along with the measured overall span of the LβH domains. Below the OatWY structure in A is the signature sequence of the hexapeptide-repeating motif (LIV)(GAED)X2(STAV)X displayed with positional indicators (i, i + 1, i + 2…) as well as the hexapeptide sequence motifs that define the OatWY LβH domain. Sheets and loops composed of the LβH domain are represented as S1–3 and L1–3, respectively. Sheets (S1–S3) are boxed with a yellow color, and the hexapeptide sequences that obey the rule ((LIV)(GAED)X2(STAV)X) are boxed with solid black rectangles, and those that do not with dashed red rectangles. The defining hexapeptide repeating motifs of each of the other acetyltransferases are also represented in a sequence table below.