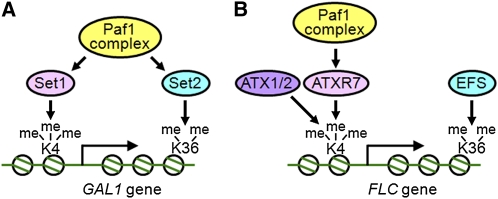
Figure 9.
Schematic Model of Transcriptional Activation through Histone Modifications.
Summary of the relationship among the factors involved in the transcriptional activation of the yeast GAL1 gene ([A]; from Hampsey and Reinberg, 2003) and the FLC gene (B). Circles, histone octamers; K4, Lys 4 of histone H3 (H3K4); K36, Lys 36 of histone H3 (H3K36); me, methyl groups on Lys residues.
(A) In yeast, the Paf1 complex is required for the recruitment of both Set1 and Set2, which are H3K4 and H3K36 methylases, respectively. Both trimethylation of H3K4 (H3K4me3) and dimethylation of H3K36 (H3K36me2) are involved in the transcriptional activation of certain genes including GAL1.
(B) In Arabidopsis, H3K4me3 and H3K36me2 are also essential for the transcriptional activation of FLC. Both Set1-class (ATXR7) and Trx-class (ATX1/2) H3K4 methylases are required for full H3K4 methylation and, thus, FLC activation. The Paf1 complex (including ELF7 and VIP4) is required for the function of ATXR7 and possibly ATX1/2, but not EFS (a Set2 ortholog).
[See online article for color version of this figure.]