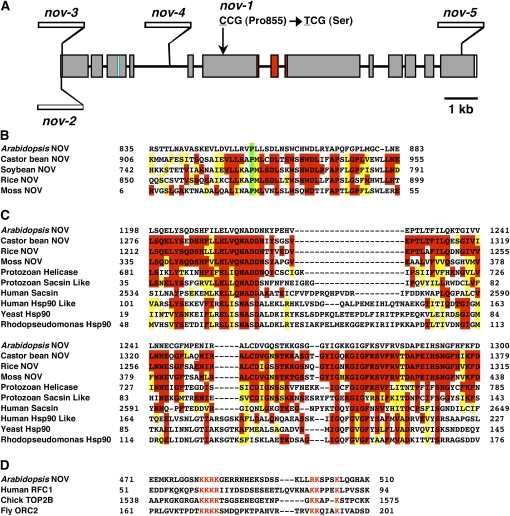
Figure 8.
Schematic Structure of NOV Gene and Some Features of NOV Protein.
(A) NOV gene is composed of 13 exons and 12 introns. The missense mutation site in nov-1 is schematically indicated by an arrow. Sites of T-DNA insertions in nov-2, -3, -4, and -5 are shown. Regions coding the putative NLS and the putative GHKL-type ATPase domain are marked in blue and in red, respectively.
(B) Alignment of protein sequences around the 855th Pro residues (marked in green) of NOV homologs. The Pro residue is conserved among NOV homologs in Arabidopsis, castor bean, soybean, rice (O. sativa Japonica group), and moss.
(C) Comparison of the putative GHKL-type ATPase domain in plant NOV homologs, Cryptosporidium parvum superfamily I helicase (Protozoan Helicase), sacsin-like protein (Protozoan Sacsin Like), human sacsin (Human Sacsin), Hsp90-like TNF receptor-associated protein 1 (Human Hsp90 Like), yeast Hsp90 (Yeast Hsp90), and R. palustris Hsp90 (Rhodopseudomonas Hsp90). In (B) and (C), residues of amino acids identical and similar to those of Arabidopsis NOV are marked in red and in yellow, respectively.
(D) The putative NLS in NOV is compared with those that are experimentally confirmed to be a NLS in human replication factor C subunit 1 (RFC1), chicks DNA topoisomerase 2-β (TOP2B), and Drosophila origin recognition complex subunit 2 (ORC2). Amino acid residues marked in red are core sequences of K[RK]{3,5}x{11,18}[RK]Kx{2,3}K motif, where K[RK]{3,5} represents for K followed by 3 to 5 of R or K, x{11,18} for 11 to 18 of any amino acid residue, [RK]K for R or K followed by K and x{2,3}K for two to three of any amino acid residues followed by K.