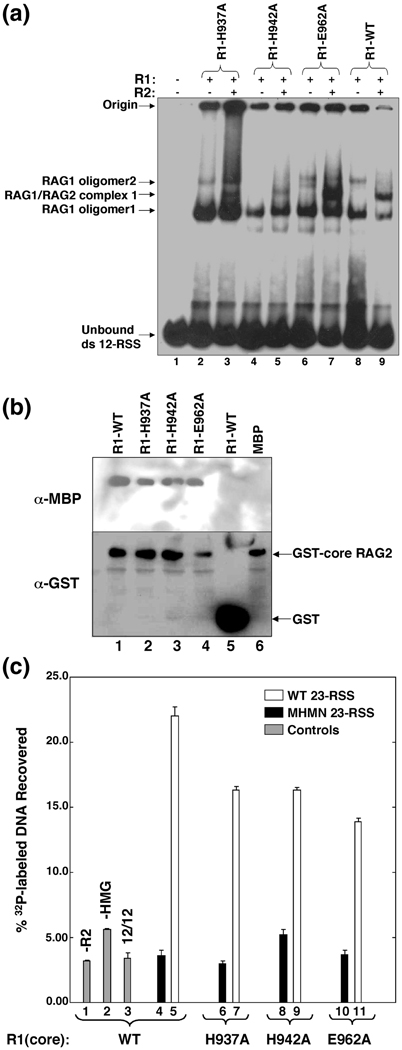
Figure 6.
The effect of mutating the zinc-binding ligands H937 and H942 on macromolecular interactions mediated by RAG1. (A) Electrophoretic mobility shift assays of mR1(core) proteins (0.04 µM) and gR2(core) (0.005 µM) combined with 1 nM 32P-labeled 12-RSS. Each lane contained 32P-labeled 12-RSS and mR1(core) and gR2(core) proteins as indicated. The mR1(core) protein used in each lane is labeled above each lane. The shifted bands in lanes containing only mR1(core) correspond to two oligomeric forms of the protein complexed with the radiolabeled 12-RSS and are labeled as RAG1 oligomer1 and oligomer2. The supershifted bands in lanes that contain both mR1(core) and gR2(core) are designated RAG1/RAG2 complex 1. The origin and unbound 32P-labeled 12-RSS are labeled accordingly. (B) Interaction between mR1(core) and gR2(core) proteins. gR2(core) was bound to gluthathione-Sepharose resin in the presence of mR1(core) proteins (WT, H937A, H942A, or E962A) or mbp. After elution from the resin, the proteins were resolved by SDS-PAGE and detected by Western blots. (C) Formation of the paired complex. In each sample (unless labeled otherwise), mR1(core) (WT or mutants) (0.2 µM) and gR2(core) (0.2 µM) proteins, and 0.3 µM HMGB1 were incubated with 10 nM biotinylated WT 12-RSS and 1 nM 32P-labeled RSS. The protein-DNA complexes were captured by streptavidin-coated paramagnetic particles as described in Materials and Methods. The bars in the graph represent the % 32P-labeled RSS captured. The mR1(core) protein used in each sample is labeled beneath the graph. Control samples are in lanes 1–3 (gray bars) and either lacked gR2(core) or HMGB1 (lanes 1 and 2, respectively), or used a 32P-labeled WT 12-RSS in place of the 23-RSS (lane 3). White bars represent samples containing 32P-labeled WT 23-RSS. Black bars represent samples containing 32P-labeled MHMN 23-RSS (where MHMN refers to a 10 RSS with mutated nonamer and heptamer sequences as described in Materials and Methods).Results are from n=4 experiments per sample.