Abstract
Cytochrome P450 2E1, gene symbol CYP2E1, is one of a family of enzymes with a central role in activating and detoxifying xenobiotics and endogenous compounds. Genetic variation at this gene has been reported in different human populations, and some association studies have reported increased risk for cancers and other diseases. To the best of our knowledge, multi-SNP haplotypes and linkage disequilibrium (LD) have not been systematically studied for CYP2E1 in multiple populations. Haplotypes can greatly increase the power both to identify patterns of genetic variation relevant for gene expression as well as to detect disease-related susceptibility mutations. We present frequency and LD data and analyses for 11 polymorphisms and their haplotypes that we have studied on over 2,600 individuals from 50 human population samples representing the major geographical regions of the world. The diverse patterns of haplotype variation found in the different populations we have studied show that ethnicity may be an important variable helping to explain inconsistencies that have been reported by association studies. More studies clearly are needed of the variants we have studied, especially those in the 5′ region, such as the VNTR, as well as studies of additional polymorphisms known for this gene to establish evidence relating any systematic differences in gene expression that exist to the haplotypes at this gene.
Keywords: CYP2E1, Cytochrome P450, SNP, haplotype, linkage disequilibrium, random genetic drift
Introduction
Haplotype diversity is a key to understanding population evolution as well as disease evolution. Heterogeneity in both Linkage Disequilibrium (LD) and haplotype frequencies across the genome have been observed among large numbers of diverse ethnic populations in several studies 1–4. Earlier studies from our laboratory have shown that haplotype and LD patterns at different genes associated with diseases vary widely across different populations of the world 2, 5–7. Studies on different genes associated with disease that included the CEPH diversity panel have also shown widely varying haplotype patterns 8–9. These earlier studies demonstrate the importance of studying the variation patterns in multiple populations representing different regions of the world for genes that have been associated with disease.
Cytochrome P450 2E1 (CYP2E1) is a member of the cytochrome P450 multifamily of enzymes that play a central role in activating and detoxifying a wide variety of xenobiotics as well as endogenous compounds. Several drug effects have been identified. The anti-fungal drug miconazale has been found10 to inhibit CYP2E1 enzyme activity. Peterson et al.11 have discussed the complex role CYP2E1 appears to play in the pharmacologic interaction of ciprofloxacin and pentoxifylline; genetic variation in CYP2E1 function may thus have complex secondary consequences. The review by Gonzalez and Yu12 summarizes the evidence for the important role that genetic variation in the CYP2E1 enzyme plays in the susceptibility of patients to hepatitis induced by anti-tuberculosis drug therapy. The PharmGKB database has links to publications showing the relationship to alcohol-related liver diseases and also reports drug response studies involving CYP2E1 for acetaminophen, alcohol, ethanol, geldanamycin, and xenobiotics.
Considerable variation in allelic distributions at CYP2E1 and of CYP2E1 enzyme activity is found among different human populations 13–16. Several polymorphic sites in the 5′-flanking and intronic region of CYP2E1 have been reported to be associated with increased risk factors for cancers and other diseases 16–20. However, no consistent results were observed in studies of the effects of these SNPs on the expression of the gene and activity of the enzyme, and on the susceptibility to diseases 21–24.
The promoter region and other regulatory variation in or near the gene will function in cis with any amino acid variation as one functional unit. Relevant variation can also include any variants that affect splicing or mRNA conformation. Thus, the haplotype encompassing all relevant variation is the relevant unit for association studies. LD may allow SNPs with no functional consequences to serve as surrogates for unknown and/or untyped variants with functional consequences. However, haplotype frequencies and LD patterns are expected to vary among populations.
Haplotypes and LD of the CYP2E1 gene region have been poorly studied. The aim of the present study has been to analyze polymorphisms across most of the CYP2E1 gene, document global ethnic variation in their allelic frequencies, and study the patterns that exist in haplotypes and LD. To those ends we present data on 11 polymorphisms, their frequencies and haplotypes in over 2,600 normal, healthy individuals from 50 population samples representing all major geographical regions of the world.
Results
A total of 2,657 mostly unrelated individuals (by self-report) were typed and analyzed for each of these polymorphisms. Allele frequencies and sample sizes for the 11 polymorphisms in all 50 populations can be found in ALFRED (http://alfred.med.yale.edu/) using the UIDs in Tables 1 and 2. Allele frequency ranges for each polymorphism are given in Figure 1, and the ancestral allele frequencies for the 10 SNPs and the most common allele frequency for the VNTR are given in Supplemental Table S1. There were no significant deviations from Hardy-Weinberg ratios. The average heterozygosities across 50 population samples and Fst values for 11 markers are shown in Supplementary Figure S1. For most markers the average heterozygosities are low, ranging from 0.035 (marker 7) to 0.283 (marker 10). Fst values vary around the mean of 0.14 for a standard set of 369 SNPs 25 but are high at markers 10 and 11, 0.254, 0.231, respectively, at the 3′ end of the gene. Only seven of the eleven markers segregate in all populations. The derived allele frequencies of SNPs at exon 4 (marker 6) and exon 6 (marker 7) are very low outside of Africa and these derived alleles are completely absent in the populations of East Asia and the Americas. The derived alleles of the upstream SNPs, except rs6413420 (marker 5), are observed in higher frequencies in Asia and the Americas than in Africa or Europe.
Table 1.
Description of 11 polymorphic markers studied in the CYP2E1 gene.
| Marker | Function | Polymorphism | dbSNP rs# | Site Location | Positiond (bp) | Base pairs to next SNP | ALFRED UIDa | ADeles | Ancestral allele |
|---|---|---|---|---|---|---|---|---|---|
| 1 | none proven | VNTR | 5′ upstream | 135,188,828 | 885b | SI01 40900 | 4 alleles | NAc | |
| 2 | C_2431875_10;Pstl | rs3813867 | 5′ upstream | 135,189,595 | 240 | SI000693S | G/G | G | |
| 3 | Rsal | rs2031920 | 5′ upstream | 135,189,835 | 703 | S1000694T | C/T | C | |
| 4 | C_15867697_1D | rs2070672 | 5′ upstream | 135,190,538 | 281 | SI001473P | G/A | A | |
| 5 | C_25594209_10 | rs641342Q | 5′ upstream | 135,190,819 | 4,846 | SI001475R | T/G | G | |
| 6 | Val179lle | C_30443971_10 | rs6413419 | exon 4 | 135,195,665 | 1,722 | SI001468T | G/A | G |
| 7 | lle321lle | C_7468401_10 | rs915909 | exon 6 | 135,197,387 | 330 | SI001476S | T/C | C |
| 8 | C_30173803_10; Mspl | rs4646976 | intron 6 | 135,197,717 | 817 | SI000692R | A/G | A | |
| 9 | Dral | rs6413432 | intron 6 | 135,198,534 | 2,593 | SI014089W SI014088V | A/T | A | |
| 10 | C_16026001_20 | rs2070676 | intron 7 | 135,201,127 | 225 | GIC | G | ||
| 11 | Phe421Phe | C_16026002_10 | rs2515641 | exon 8 | 135,201,352 | SI000174Q | TC | C |
VNTR has two common (6-, 8-repeats described by Hu el al.1999) alleles; two rare alleles observed in African samples.
“UIDs” are unique identifiers in the ALFRED database for polymorphism descriptions and allele frequencies.
Distance to next SNP (bp) from the proximal end of the VNTR
Not Available; Failed to identify ancestral allele for VNTR
NCBI Map Build 36.3
Table 2.
The 50 Population samples studied with unique identifier links to full descriptions in the ALFRED database.
| Name | Abbrev. | Location | N | Population ALFRED UID | Sample ALFRED UID |
|---|---|---|---|---|---|
| Africa | |||||
| Biaka | BIA | S.W. Central African Rep. | 70 | PO000005F | SA000005F |
| Mbuti | MBU | eastern Dem. Rep. Congo | 39 | PO000006G | SA000006G |
| Yoruba | YOR | western Nigeria | 78 | PO000036J | SA000036J |
| Ibo | IBO | southern Nigeria | 48 | PO000096P | SA000099S |
| Hausa | HAS | northern Nigeria | 39 | PO000097Q | SA000100B |
| Chagga | CGA | Kilimanjaro area, Tanzania | 45 | PO000324J | SA000487T |
| Masai | MAS | Northern Tanzania | 22 | PO000456P | SA000854R |
| Sandawe | SND | North Central. Tanzania | 40 | PO000661N | SA001773S |
| African Americans | AAM | United States | 90 | PO000098R | SA000101C |
| Ethiopian Jews | ETJ | northwestern Ethiopia ‡ | 32 | PO000015G | SA000015G |
| Somali | SOM | Somalia; refugees in Pakistan | 40 | PO000075M | SA0021380 |
| S.W. Asia, Europe | |||||
| Yemenite Jews | YMJ | Yemen ‡ | 43 | PO000085N | SA000016H |
| Druze | DRU | Israel | 106 | PO000008I | SA000047L |
| Samaritans | SAM | Israel | 41 | PO0000950 | SA000098R |
| Ashkenazi | ASH | eastern Europe ‡ | 83 | PO000038L | SA000490N |
| Adygei | ADY | Krasnodar, Caurasus Mfns | 54 | PO000017I | SA000017I |
| Chuvash | CHV | easternmost Europe near Urals | 42 | PO000327M | SA0004910 |
| Hungarians | HGR | Hungary | 92 | PO000453M | SA002023H |
| Russians | RUA | Kargopol, Archangelsk region | 34 | PO000019K | SA001530J |
| Russians | RUV | Vologda, northern Russia | 48 | PO000019K | SA000019K |
| Finns | FIN | Finland | 36 | PO000018J | SA000018J |
| Danes | DAN | Denmark | 51 | PO000007H | SA000007H |
| Irish | IRI | Ireland | 118 | PO000057M | SA000057M |
| Euro Americans | EAM | United States | 92 | PO000020C | SA000020C |
| N.W. Asia (Siberia) | |||||
| Komi Zyriane | KMZ | N.W. Asia, near Urals | 47 | PO000326L | SA000489V |
| Khanty | KTY | N.W. Asia near Urals | 50 | PO000325K | SA000488U |
| S.C. Asia | |||||
| Mohanna | MHN | Pakistan | 112 | PO000708P | SA002139P |
| Hazara | HZR | Pakistan | 58 | PO000575R | SA002140H |
| Negroid Makrani | NMK | Pakistan | 56 | PO000707O | SA002137N |
| Keralites | KER | Kerala, India ↕ | 30 | PO000672P | SA001854S |
| N.E. Asia (Siberia) | |||||
| Yakut | YAK | Sakha, N.E. Siberia | 51 | PO000011C | SA000011C |
| Pacific Islands | |||||
| Nasioi | NAS | Bougainville, Solomon Islands | 23 | PO000012D | SA000012D |
| Micronesians | MCR | Micronesia, multiple islands | 37 | PO000063J | SA000063J |
| East Asia | |||||
| Laotians | LAO | Laos | 119 | PO000671O | SA001853R |
| Cambodians | CBD | Cambodia | 25 | PO000022E | SA000022E |
| SF Chinese | CHS | southern Han, SF Bay Area | 60 | PO000009J | SA000009J |
| TW Chinese | CHT | Taiwan | 49 | PO000009J | SA000001B |
| Hakka | HKA | Taiwan | 41 | PO000003D | SA000003I |
| Koreans | KOR | Seoul, Korea | 54 | PO000030D | SA000936S |
| Japanese | JPN | Japan | 51 | PO000010B | SA000010B |
| Ami | AMI | eastern mtns, Taiwan | 40 | PO000002C | SA000002C |
| Atayal | ATL | eastern mtns, Taiwan | 42 | PO000021D | SA00002ID |
| Americas | |||||
| Cheyenne | CHY | Oklahoma, U.S.A. | 56 | PO000023F | SA000023F |
| Pima, Arizona | PMA | Arizona, United States | 51 | PO000033G | SA000025H |
| Pitna, Mexico | PMM | northern Mexico | 53 | PO000034H | SA000026I |
| Maya | MAY | central Yucatan, Mexico | 52 | PO000013E | SA000013E |
| Quechua | QUE | Peru | 22 | PO000069P | SA000069P |
| Ticuna | TIC | Amazon, Brazil | 65 | PO000027J | SA000027J |
| R. Surui | SUR | Rondonia, Amazon, Brazil | 47 | PO000014F | SA000014F |
| Karitiana | KAR | Amazon, Brazil | 57 | PO000028K | SA000028K |
Samples collected in Israel
Samples collected in U.S.A. from individuals born in Kerala
Figure 1.
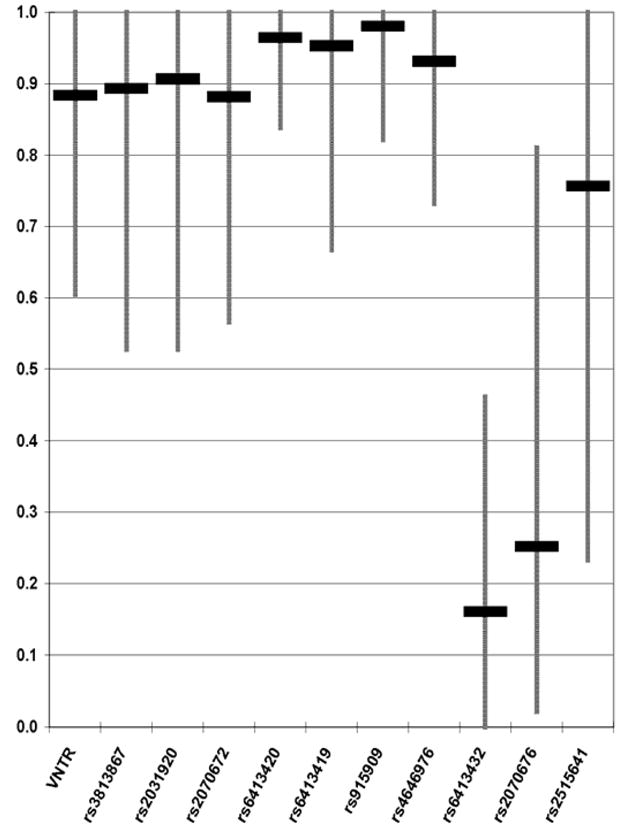
Graphical representation of the average and range of ancestral allele frequencies in 50 population samples for each of 11 markers at CYP2E1 in 50 population samples.
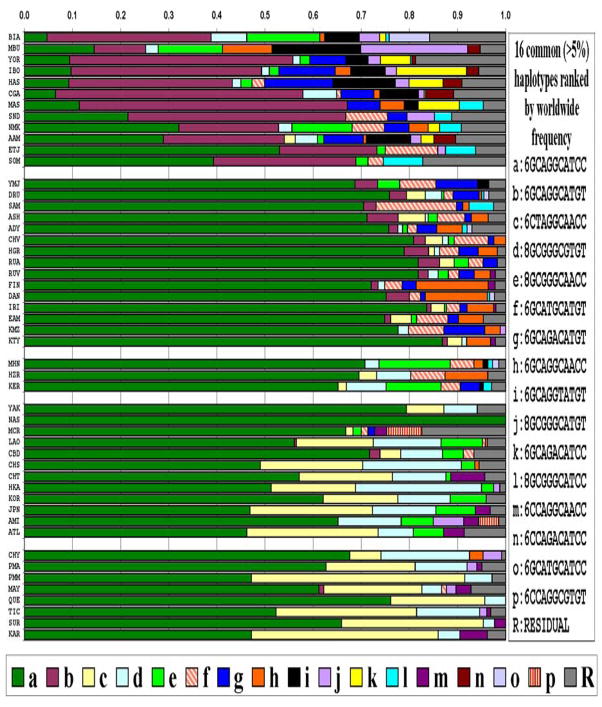
We inferred 16 common haplotypes and estimated their frequencies (Figure 2). Most of the low frequency variation in the residual class of rare haplotypes is accounted for by a relatively small number of haplotypes in the 2 to 4% frequency range. The variation in haplotype frequencies among populations gives rise to a complex pattern of LD, both pairwise and as segments with high LD, that varies among populations (Supplemental Table S2 and Supplemental Figure S2).
Figure 2.
Frequencies for the haplotypes based on 11 markers at CYP2E1 for 50 populations. For each color-coded haplotype the alleles are shown for the sites in chromosome order as numbered in Table 1. Both the full allelic description and lower-case letter codes for the haplotypes are given. For each population the proportional length of each color bar represents the frequency of the respective haplotype. All haplotypes that have frequencies less than 5% in all the populations studied are grouped into the residual (gray bar) class. The ancestral haplotype, GCAGGCAAGC, for markers 2 through 11, is not found at common frequencies in any of the 50 populations studied.
Haplotype diversity is much higher in Africa (with 6–10 common haplotypes) than outside of Africa (with about 1–6 common haplotypes). The most common 11-MARKER haplotype, 6GCAGGCATCC (dark green in Figure 2), is very frequent in all populations outside of Africa and in Ethiopia, but not in other populations of Africa. Two haplotypes, 6CTAGGCAACC (light yellow) and 8GCGGGCGTGT (light blue), are not seen in African populations and rarely seen (<5%) in European populations, but are more frequent in most East Asian (0.0 – 0.273 and 0.073 – 0.262) and Native American (0.065 – 0.443 and 0.023 – 0.184) populations.
In order to understand the evolution of the haplotypes we estimated haplotypes with fewer SNPs across shorter segments of the gene. We identified three core regions that have evolved common haplotypes solely by accumulation of mutations from the ancestral core haplotype. These cores involve markers 1 through 5 (core A), markers 6 through 9 (core B), and markers 10 and 11 (core C) (Figure 3). No recurrent mutations are required to explain all of these core haplotypes. The full 11-marker haplotypes can be explained by combinations of the haplotypes of the three cores (Figure 4 and Supplemental Table S3). These combinations have arisen by accumulation of mutations (as depicted in Figure 3) and historical crossovers. It is difficult to be certain of orders of all events, mutations and crossovers, when the three cores are considered together, in part because other combinations that could have been intermediate now are either absent or exist among the rare haplotypes.
Figure 3.
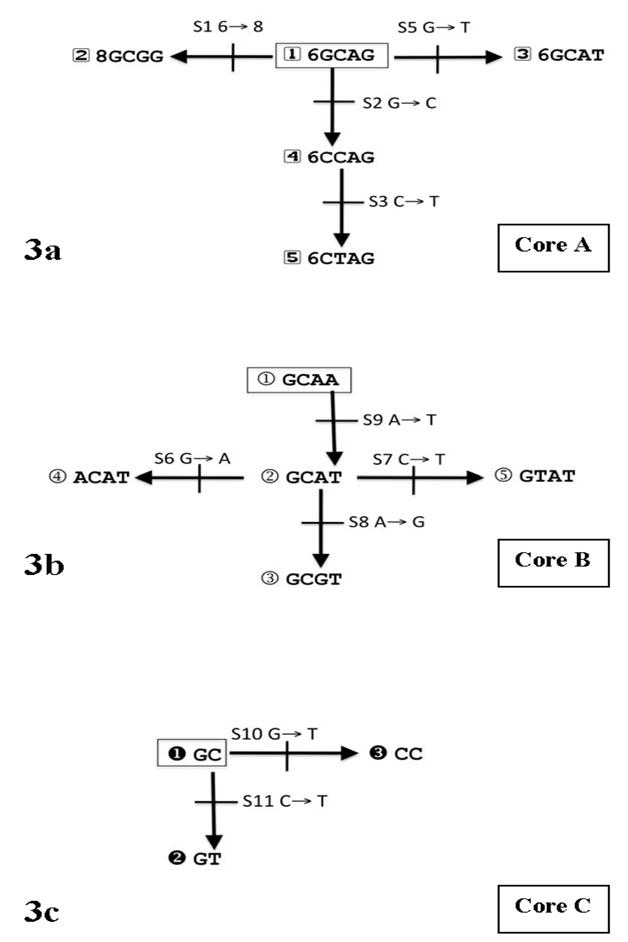
The evolutionary relationships among haplotypes of three core segments of CYP2E1. In all cases the schema starts with the ancestral human haplotype and gives the pattern of mutational accumulation for that core. A: Core A comprised of markers 1 through 5. B: Core B comprised of markers 6 through 9. C: Core C comprised of markers 10 and 11.
Figure 4.
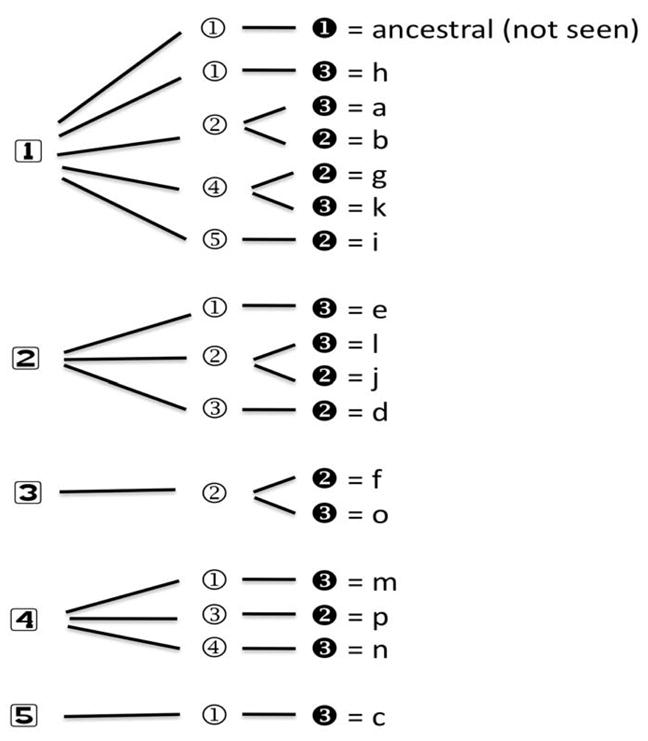
The composition of the 11 full haplotypes in terms of combinations of individual core haplotypes. The core haplotypes are numbered as in Figure 3 with core A on the left, core B in the center, and core C on the right. The lower case letters for the full haplotypes correspond to those in Figure 2. (See also Supplemental Table S3.)
In contrast to the global frequency patterns of the whole 11-marker haplotypes, the individual core haplotypes show different global patterns (Supplemental Figures 3, 4, & 5). Core A haplotypes show greater frequency similarity between African and European populations than between European and both East Asian and Native American populations. One core A haplotype, 6GCAG (#1 in Figure 3A), exists at frequencies of 56% to 95% in the Africans and Europeans. Another core A haplotype, 6CTAG (#5 in Figure 3A), is not seen in Africans, is rare in Europeans, but is frequent in East Asian and Native American populations. To the degree these 5′ markers encompass the major regulatory regions, it is possible that East Asians and Native Americans may have a common derived variant in regulation that is very uncommon to absent in the rest of the world.
Discussion
We are unaware of any publications of the CYP2E1 gene that have included all of the polymorphisms that we present here. Certainly, none of these markers has been studied previously on such a large and ethnically diverse set of individuals. This study is an explicit example of the type of global perspective on pharmacogenetic variation within and among populations discussed in an editorial by Marsh26. Even this dataset does not probe the full extent of the genetic diversity of this small segment of DNA. Public databases report multiple additional polymorphisms across the gene (including 5′ and 3′ UTRs).
We cannot precisely relate the 16 common haplotypes (Figure 2) we have observed to the standard CYP2E1 allelic designations in the “cypalleles” web site (http://www.cypalleles.ki.se/cyp2e1.htm) because, from a genetic transmission perspective, each of the haplotypes we report is an allele and the “cypalleles” web site does not give full haplotype specifications for the allele designations they summarize, precluding a strict comparison. Moreover, we have not included SNPs with rare or uncommon variants that have not been studied widely. To distinguish the haplotypes we have identified from those in the “cypalleles” nomenclature, we have used letter designations rather than numbers in Figures 2 and 4 and Supplemental Table S3. As an example of the difficulty of establishing precise correspondences, the mutation G→A at marker 6 (corresponding to 179 Val→Ile) defines core B haplotype 4 (Figure 3B) and appears to represent one mutational event. That core B haplotype exists in two combinations with core A haplotypes and two combinations with core C haplotypes for a total of three 11-marker haplotypes: g, k, and n (Figures 2 & 4). All three of these haplotypes correspond to allele CYP2E1*4 in the “cypalleles” nomenclature. We expect the haplotypes encompassing the gene to become more complex as more SNPs and rare variants are included in an even more comprehensive study of the gene.
In addition to multiple SNPs across this gene, copy number variation (CNV) encompassing CYP2E1 has been reported27–27. Our typing methods are not designed to detect CNVs but we can exclude any common occurrence in our samples because there is no significant deviation from HW ratios in any of the populations.
We have studied the allele, haplotype, and LD variation patterns for 11 polymorphisms in 50 populations from different geographical regions of the world across the CYP2E1 gene and have shown that there are large differences in these patterns worldwide. The haplotypes were useful in inferring recombination events in the recent evolution of the gene. The current study focuses attention on the core haplotype lineages that appear to have involved no recombination and on the combinations that have arisen because of historical recombinations. These cores and their combinations provide the framework for future expression studies. Depending upon when in one of the evolutionary lineages a functional variant arose, we would expect it to either define a new sublineage or be inherited into the descendant haplotypes in Figure 3. Thus, the evolutionary lineages may explain multiple haplotypes (alleles) having similar functional properties, even if the causative variant has not yet been identified. Other SNPs within the molecular extent of the region spanned will likely fall within this framework; some might refine the locations of the inferred historical crossovers.
Supplementary information is available at the The Pharmacogenomics Journal’s website.
Materials and Methods
Samples studied
DNA was purified from lymphoblastoid cell lines from 2657 healthy adults from 50 populations from around the world (Table 2). Population membership was designated by the subjects and all blood samples were obtained with individual informed consent following protocols approved by the Institutional Review Boards at Yale University School of Medicine, at the University of Karachi, and at multiple other relevant institutions in countries where samples were collected. The average population sample size is 53 individuals.
Markers studied
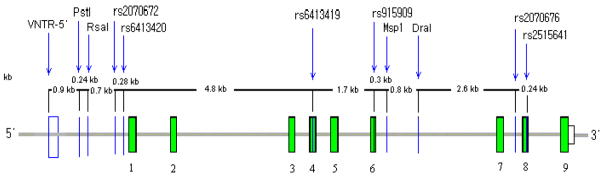
We studied ten SNPs and one VNTR across 13.9 kb that encompasses the 5′ region of the CYP2E1 gene and almost the entire coding region (Figure 5). We typed the VNTR and four SNPs in the upstream region, three SNPs in the coding regions, and three SNPs in the intronic regions of the gene (Table 1). The markers are referred to by their numeric position (1–11) in Table 1. The SNPs are all diallelic and the VNTR is essentially diallelic, as initially described29. Two other very rare VNTR alleles have been seen in some African populations in the course of this study (data not shown); they were excluded from the haplotype analyses.
Figure 5.
Map of CYP2E1 on chromosome 10 and the markers typed. The filled boxes represent the exons of the gene; the number below each box is the exon number. The vertical lines represent positions of the markers studied.
Typing methods
The samples were typed by TaqMan assays (markers 2, 4–8, 10, and 11), by fragment length analysis on agarose gels (marker 1) after PCR, and by restriction fragment length after enzyme digestion of the PCR products for markers 3 (RsaI) and 9 (DraI).
Determining Ancestral Alleles
For each allele, the ancestral state in humans was determined by inference from the allele present in several other primate species. The ancestral allele of the VNTR (marker 1) could not be determined but by inference is 6.
Statistical methods
Allele frequencies of the VNTR and SNPs were calculated by gene counting assuming co-dominant inheritance. All the sites were also tested for Hardy-Weinberg ratios by chi-square test and/or exact test. Expected heterozygosities were estimated as 1-Σpi2. Haplotype frequencies were estimated by the EM algorithm using HAPLO 30. Haplotypes with estimated frequencies of less than 5% in each of the population samples go into the residual class. The 5% threshold is a reasonble boundary for determining what are the common and rare haplotypes given the sample sizes in this study. While some estimated haplotypes below the 5% threshold have very clear evidence of occurrence, the standard errors on estimated frequencies increase along with some erroneous inferences due to the small number of observations available in the rare zone and the fact that the LD levels between sites vary. Pair-wise LD estimates were done as r2 [refs. 31, 32] with significance levels determined by a permutation test33. Comparative plots of LD for all the populations were done using HAPLOT 34.
Supplementary Material
Acknowledgments
This work was funded, in part, by National Institute of Health grants GM057672 and AA009379 to Kenneth K. Kidd. None of the authors has any conflicts of interest related to the data presented here. We thank the many colleagues who helped us assemble the population samples. Special thanks are due the many hundreds of individuals from these populations who volunteered to give blood samples for studies such as this.
Abbreviations
- CYP2E1
cytochrome P450, family 2, subfamily E, polypeptide 1
- LD
linkage disequilibrium
- PCR
polymerase chain reaction
- SNP
single nucleotide polymorphism
- UID
unique identifier
- VNTR
variable number of tandem repeats
Footnotes
Duality of Interest
None declared.
Electronic Databases Cited
ALFRED, The ALlele FREquency Databse; http://alfred.med.yale.edu
PharmGKB, The Pharmacogenetics and Pharmacogenomics Knowledge Base, http://www.PharmGKB.org
References
- 1.Kidd KK, Morar B, Castiglione CM, Zhao H, Pakstis AJ, Speed WC, et al. A global survey of haplotype frequencies and linkage disequilibrium at the DRD2 locus. Hum Genet. 1998;103:211–227. doi: 10.1007/s004390050809. [DOI] [PubMed] [Google Scholar]
- 2.Kidd JR, Pakstis AJ, Zhao H, Lu RB, Okonofua FE, Odunsi A, et al. Haplotypes and linkage disequilibrium at the phenylalanine hydroxylase locus, PAH, in a global representation of populations. Am J Hum Genet. 2000;66:1882–1899. doi: 10.1086/302952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sawyer SL, Mukherjee N, Pakstis AJ, Feuk L, Kidd JR, Brookes AJ, Kidd KK. Linkage disequilibrium patterns vary substantially among populations. Eur J Hum Genet. 2005;13:677–686. doi: 10.1038/sj.ejhg.5201368. [DOI] [PubMed] [Google Scholar]
- 4.Frisse L, Hudson RR, Bartoszewicz A, Wall JD, Donfack J, Di Rienzo A. Gene conversion and different population histories may explain the contrast between polymorphism and linkage disequilibrium levels. Am J Hum Genet. 2001;69:831–843. doi: 10.1086/323612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mukherjee N, Kidd KK, Pakstis AJ, Speed WC, Li H, Tarnok Z, Barta C, Kajuna SLB, Kidd JR. The complex global pattern of genetic variation and linkage disequilibrium at Catechol-O-methyl transferase (COMT) Molecular Psychiatry. doi: 10.1038/mp.2008.64. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han Y, Gu S, Oota H, Osier MV, Pakstis AJ, Speed WC, Kidd JR, Kidd KK. Evidence of positive selection on a class I ADH locus. Am J Hum Genet. 2007;80:441–456. doi: 10.1086/512485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tishkoff SA, Goldman A, Calafell F, Speed WC, Deinard AS, Bonne-Tamir B, et al. A global haplotype analysis of the myotonic dystrophy locus: implications for the evolution of modern humans and for the origin of myotonic dystrophy mutations. Am J Hum Genet. 1998;62:1389–1402. doi: 10.1086/301861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evans DM, Cardon LR. Guidelines for genotyping in genomewide linkage studies: single-nucleotide-polymorphism maps versus microsatellite maps. Am J Hum Genet. 2004;75:687–692. doi: 10.1086/424696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gardner M, González-Neira A, Lao O, Calafell F, Bertranpetit J, Comas D. Extreme population differences across Neuregulin 1 gene, with implications for association studies. Mol Psychiatry. 2006;11:66–75. doi: 10.1038/sj.mp.4001749. [DOI] [PubMed] [Google Scholar]
- 10.Niwa T, Inoue-Yamamoto S, Shiraga T, Takagi A. Effect of antifungal drugs on Cytochrome P450 (CYP) 1A2, CYP2D6, and CYP2E1 activities in human liver microsomes. Biol Pharm Bull. 2005;28:1813–1816. doi: 10.1248/bpb.28.1813. [DOI] [PubMed] [Google Scholar]
- 11.Peterson TC, Peterson MR, Wornell PA, Blanchard MG, Gonzalez FJ. Role of CYP1A2 and CYP2E1 in the pentoxifylline ciprofloxacin drug interaction. Biochemical Pharmacology. 2004;68:395–402. doi: 10.1016/j.bcp.2004.03.035. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez FJ, Yu AM. Cytochrome P450 and xenobiotic receptor humanized mice. Ann Rev Pharmacol Toxicol. 2006;46:41–64. doi: 10.1146/annurev.pharmtox.45.120403.100007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayashi S, Watanabe J, Nakachi K, Kawajiri K. Genetic linkage of lung cancer-associated MspI polymorphisms with amino acid replacement in the heme binding region of the human cytochrome P450IA1 gene. J Biochem (Tokyo) 1991;110:407–411. doi: 10.1093/oxfordjournals.jbchem.a123594. [DOI] [PubMed] [Google Scholar]
- 14.Kim RB, Yamazaki H, Chiba K, O’Shea D, Mimura M, Guengerich FP, et al. In vivo and in vitro characterization of CYP2E1 activity in Japanese and Caucasians. J Pharmacol Exp Ther. 1996;279:4–11. [PubMed] [Google Scholar]
- 15.Garte S, Gaspari L, Alexandrie AK, Ambrosone C, Autrup H, Autrup JL, et al. Metabolic gene polymorphism frequencies in control populations. Cancer Epidemiol Biomarkers Prev. 2001;10:1239–1248. [PubMed] [Google Scholar]
- 16.Danko IM, Chaschin NA. Association of CYP2E1 gene polymorphism with predisposition to cancer development. Experimental Oncology. 2005;27:248–256. [PubMed] [Google Scholar]
- 17.Song BJ. Ethanol-inducible cytochrome P450 (CYP2E1): biochemistry, molecular biology and clinical relevance: 1996 update. Alcohol Clin Exp Res. 1996;20(Suppl):138A–146A. doi: 10.1111/j.1530-0277.1996.tb01764.x. [DOI] [PubMed] [Google Scholar]
- 18.Iwahashi HK, Ameno S, Ameno K, Okada N, Kinoshita H, Sakae Y, et al. Relationship between alcoholism and CYP2E1 C/D polymorphism. Neuropsychobiology. 1998;38:218–221. doi: 10.1159/000026544. [DOI] [PubMed] [Google Scholar]
- 19.Nguyen TT, Murphy NP, Austin CM. Amplification of multiple copies of mitochondrial Cytochrome b gene fragments in the Australian freshwater crayfish, Cherax destructor Clark (Parastacidae: Decapoda) Anim Genet. 2002;33:304–308. doi: 10.1046/j.1365-2052.2002.00867.x. [DOI] [PubMed] [Google Scholar]
- 20.Howard LA, Ahluwalia JS, Lin SK, Sellers EM, Tyndale RF. CYP2E1*1D regulatory polymorphism: association with alcohol and nicotine dependence. Pharmacogenetics. 2003;13(6):321–8. doi: 10.1097/01.fpc.0000054090.48725.a2. Erratum in: Pharmacogenetics 2003;13:441–442. [DOI] [PubMed] [Google Scholar]
- 21.Lee HS, Yoon JH, Kamimura S, Iwata K, Wataname H, Kim CY. Lack of association of cytochrome P4502 E1 genetic polymorphisms with the risk of human hepatocellular carcinoma. Int J Cancer. 1997;71:737–740. doi: 10.1002/(sici)1097-0215(19970529)71:5<737::aid-ijc8>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 22.Morita S, Yano M, Shiozaki H, Tsujinaka T, Ebisui C, Morimoto T, et al. CYP1A1, CYP2E1 and GSTM1 polymorphisms are not associated with susceptibility to squamous-cell carcinoma of the esophagus. Int J Cancer. 1997;71:192–195. doi: 10.1002/(sici)1097-0215(19970410)71:2<192::aid-ijc11>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 23.Powell H, Kitteringham NR, Pirmohamed M, Smith DA, Park BK. Expression of cytochrome P4502E1 in human liver: assessment by mRNA, genotype and phenotype. Pharmacogenetics. 1998;8(5):411–21. doi: 10.1097/00008571-199810000-00006. [DOI] [PubMed] [Google Scholar]
- 24.Wong NA, Rae F, Simpson KJ, Murray GD, Harrison DJ. Genetic polymorphisms of cytochrome p4502E1 and susceptibility to alcoholic liver disease and hepatocellular carcinoma in a white population: a study and literature review, including meta-analysis. Mol Pathol. 2000;53(2):88–93. doi: 10.1136/mp.53.2.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kidd KK, Pakstis AJ, Speed WC, Kidd JR. Understanding human DNA sequence variation. J Hered. 2004;95(5):406–20. doi: 10.1093/jhered/esh060. [DOI] [PubMed] [Google Scholar]
- 26.Marsh S. Pharmacogenetics: global clinical markers. Pharmacogenomics. 2008;9(4):371–373. doi: 10.2217/14622416.9.4.371. [DOI] [PubMed] [Google Scholar]
- 27.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–54. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF, Hakonarson H, Bucan M. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–74. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu Y, Hakkola J, Oscarson M, Ingelman-Sundberg M. Structural and functional characterization of the 5′-flanking region of the rat and human cytochrome P450 2E1 genes: identification of a polymorphic repeat in the human gene. Biochem Biophys Res Commun. 1999;263:286–293. doi: 10.1006/bbrc.1999.1362. [DOI] [PubMed] [Google Scholar]
- 30.Hawley ME, Kidd KK. HAPLO: a program using the EM algorithm to estimate the frequencies of multi-site haplotypes. J Hered. 1995;86:409–411. doi: 10.1093/oxfordjournals.jhered.a111613. [DOI] [PubMed] [Google Scholar]
- 31.Devlin B, Risch N. A comparison of linkage disequilibrium measures for fine-scale mapping. Genomics. 1995;29:311–322. doi: 10.1006/geno.1995.9003. [DOI] [PubMed] [Google Scholar]
- 32.Lewontin RC. The Interaction of Selection and Linkage. Ii Optimum Models Genetics. 1964;50:757–782. doi: 10.1093/genetics/50.4.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao H, Pakstis AJ, Kidd JR, Kidd KK. Assessing linkage disequilibrium in a complex genetic system. I Overall deviation from random association. Ann Hum Genet. 1999;63:167–179. doi: 10.1046/j.1469-1809.1999.6320167.x. [DOI] [PubMed] [Google Scholar]
- 34.Gu S, Pakstis AJ, Kidd KK. HAPLOT: a graphical comparison of haplotype blocks, tagSNP sets and SNP variation for multiple populations. Bioinformatics. 2005;21:39938–3939. doi: 10.1093/bioinformatics/bti649. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.