Abstract
Transposon mini-Tn7 vectors insert into the chromosome of several Gram-negative bacteria in a site-specific manner. Here, we demonstrated the application of mini-Tn7 as single copy site-specific integration vector system for Xanthomonas campestris pv. campestris. The transposition of the mini-Tn7 into the bacterial genome was detected at a Tn7 attachment (attTn7) site located downstream of glmS1. Furthermore, using a newly constructed vector pBBR1FLP2 containing the FLP recombinase for site-specific excision of the sequence between the FLP recognition target (FRT) sites, and a sacB counter selection marker, an unmarked mini-Tn7 insertion mutant was created. Mini-Tn7 insertion did not affect bacterial virulence on the tested plant. The mini-Tn7 and FLP-FRT systems also work well in X. oryzae pv. oryzae.
Keywords: Single copy number gene cloning, mini-Tn7, unmarked mutation, Xanthomonas, virulence
Introduction
Transposons are mobile genetic elements that can transpose within the same, or other, genomes. Transposon Tn7 and its derivatives have been shown to exert a high frequency of transposition into specific target sites on the chromosome of several bacteria (Arciszewska et al., 1989). Integration of the transposable element requires the proteins encoded by Tn7 genes (tnsABCDE) and can occur using two different mechanisms (Waddell & Craig, 1989). TnsABC + TnsD promote transposition into a specific Tn7 attachment site, attTn7, with a high frequency, while TnsABC + TnsE promote insertion into non-attTn7 sites (Peters & Craig, 2001). Generally, attTn7 is located immediately downstream of glmS, a gene encoding glucosamine-fructose-6-phosphate aminotransferase, an enzyme that catalyzes the formation of glucosamine 6-phosphate. Most bacteria possess a single glmS gene and therefore a single attTn7 site. However, multiple glmS genes and attTn7 sites have been reported in some bacteria including Burkholderia spp. (Choi et al., 2006; Choi et al., 2008). Moreover, non-glmS-linked attTn7 sites have also been identified in Proteus mirabilis (Choi & Schweizer, 2006). Mini-Tn7 vectors have been developed and proven to be useful for integration of a cloned gene and genetic analysis of several Gram-negative bacteria (Choi & Schweizer, 2006; Choi & Schweizer, 2006; Choi et al., 2006; Choi et al., 2005; Peters & Craig, 2001; Waddell & Craig, 1989). These mini-Tn7-based vectors are a valuable genetic tool for examining gene complementation and expression analysis at either the transcriptional or translational level.
Xanthomonas spp. are Gram-negative aerobic bacteria that are soil and plant pathogens that cause destructive diseases in many economically important crops, including rice, citrus, cabbage, cauliflower and radish. Xanthomonas campestris is also the producer of industrially important Xanthan gum. The development of novel genetic tools to study these bacterial phytopathogens and industrially important bacteria could lead to a better understanding of the underlying pathophysiology, improve the treatment strategies and improve production of an industrial material Xanthan gum. However, one of the important problems for molecular characterization of genes in Xanthomonas is lacking of vectors for specific integration of genes into bacterial chromosome either for single copy gene complementation or in vivo promoter analysis. In this communication, we demonstrate the successful application of the mini-Tn7 vector system for integration of genes in X. campestris as well as X. oryzae. Although these bacterial species contain two glmS genes, insertion of the transposon was only detected at the glmS1-associated attTn7 site, and this insertion did not affect bacterial virulence. A new vector containing FLP recombinase that facilitates excision of FRT sites a sacB a counter section marker are useful for making unmarked mutation in Xanthomonas.
Materials and methods
Bacterial strain and culture conditions
Xanthomonas strains were grown in Silva-Buddenhagen (SB) medium (Chauvatcharin et al., 2005) at 28°C with continuous shaking. Bacteria containing genetic elements expressing sacB were cultivated in modified SB in which glucose was used instead of sucrose; sacB-deficient, sucrose-resistant clones were then selected on SB medium supplemented with 5% sucrose. Antibiotic concentrations used were 5 µg−1 ml gentamicin (Gm) and 300 µg−1 ml carbenicillin (Cb).
Plasmids
pUC18-mini-Tn7T-Gm, pUC18-mini-Tn7T-Gm-lacZ and the pTNS2 helper plasmid have been described previously (Choi & Schweizer, 2006; Choi et al., 2006; Choi et al., 2005). The FLP-mediated excision plasmid, pBBR1-FLP2 was constructed by cloning the 5-kb Acc65I-SphI (blunt) fragment of pFLP2 (Hoang et al., 1998), which contained the cI857-FLP-sacB genes, into pBBR1MCS-4 (Kovach et al., 1995) digested with Acc65I and SmaI. The complete sequence of pBBR1-FLP2 was deposited to the GenBank databases under the accession number FJ797950.
pTn7T-PahpC::lacZ was constructed by PCR amplification of 270-bp ahpC (xcc0834) promoter fragment using two specific primers (BT2305: 5’TTGCCGTTGTGGTACGCG3’ and BT2306: 5’AGCCTCAGACATGCGGCA3’) designed from the Xcc genome sequence (da Silva et al., 2002). The PCR product was cloned into pDrive cloning vector (Qaigen, Germany) prior to the subcloning of the XhoI and Acc65I fragment into pUC18-mini-Tn7T-Gm-lacZ digested with the same restriction enzymes to generate pTn7T-PahpC::lacZ.
Molecular genetic techniques
Molecular genetic techniques including genomic and plasmid DNA preparations, polymerase chain reaction (PCR), restriction endonuclease digestion, DNA ligation, transformation in Escherichia coli, bacterial crude lysate preparation, gel electrophoresis and Southern blotting analysis were performed using standard protocols (Sambrook et al., 1989). Transformation of X. campestris pv. phaseoli (Xcc) was performed by electroporation, as previously described (Mongkolsuk et al., 1996). DNA sequences were determined on an ABI 310 automated DNA sequencer (Applied Biosystems, USA).
Virulence test of X. campestris pv. campestris
The virulence of Xcc was determined on Chinese radish (Raphanus sativus), a compatible host plant, using leaf-clipping methods (Dow et al., 2003) with some modifications. Overnight cultures of Xanthomonas strains in AB minimal medium (Chilton et al., 1974) supplemented with 0.1 % (w/v) casamino acid were diluted to an optical density at 600 nm (OD600) of 1.0 in fresh AB medium. Three leaves per plant were inoculated by leaf clipping, and five leaves were inoculated for each bacterial strain. The lesion lengths were measured at 14 days post-inoculation. Experiments were done in triplicate. The difference between strains was analyzed by t-test, and P-values less than 0.05 were considered to be significantly different.
β-galactosidase assay
Crude bacterial lysates were prepared and protein assays were performed as previously described (Panmanee et al., 2002). The total protein concentration in the cleared lysate was determined using dye-binding method (Bio-Rad, USA). β-galactosidase assays were performed as previously described and expressed as international units defined as the amount of enzyme capable of releasing 1 µmol p-nitrophenol generated at 25°C per min (Panmanee et al., 2002).
Results and Discussion
Insertion of Mini-Tn7 vectors into the Xanthomonas chromosome
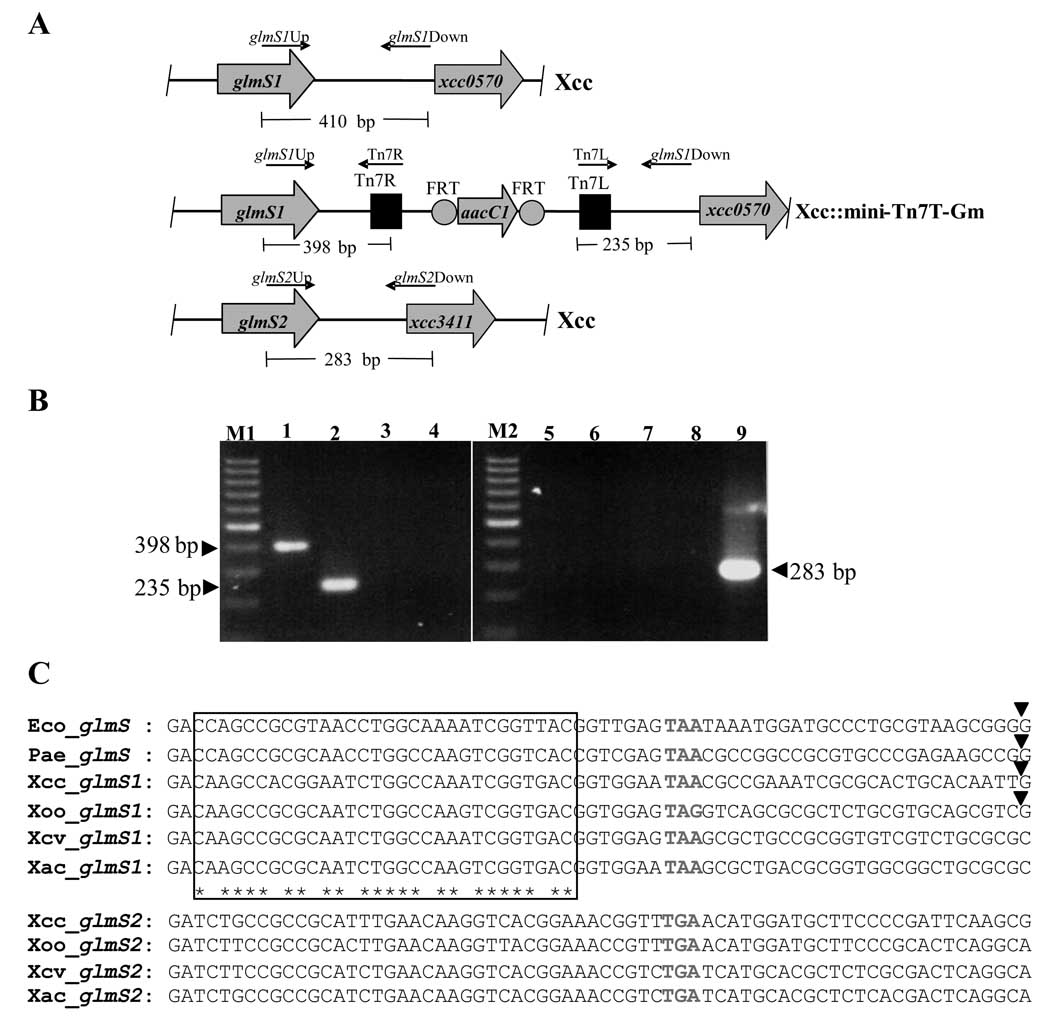
In order to examine the utility of Tn7-based genetic tools vectors for the study of Xanthomonas campestris pv. campestris (Xcc), we tested whether a mini-Tn7 vector could be used in Xcc. pUC18-mini-Tn7T-Gm and pTNS2 (the helper plasmid encoding the TnsABCD site-specific transposition pathway) were transformed into wild-type Xcc. The transformants were selected on SB plates containing 5 µg−1 ml gentamicin. The analysis of the annotated Xcc genomic sequence using the BLASTP algorithm (Altschul et al., 1997) and Pseudomonas aeruginosa glmS as the query sequence revealed that Xcc contains two putative glucosamine-fructose-6-phosphate aminotransferase genes, glmS1 (xcc0569) and glmS2 (xcc3411). Therefore, the insertion of the mini-Tn7 was verified using PCR with glmS1- or glmS2-specific forward primers and a reverse primer that located in the mini-Tn7 (Fig. 1a). The results demonstrate that the mini-Tn7-Gm was inserted downstream of glmS1 in all 37 transformants that were analyzed in this study. Fig. 1b shows the PCR products of 398 bp and 235 bp amplified from a representative transformant using glmS1Up (5’ACGACCGCCTGCTGGAAAA3’) and Tn7R (5’-CACAGCATAACTGGACTGATT3’) primers, and glmS1Down (5’-ACGGGATGGCTGCGGCTT3’) and Tn7L (5’-ATTTGCTTACGACGCTACACC3’) primers, respectively. Amplification with glmS1Up and Tn7L or glmS1Down and Tn7R primer pairs resulted in no PCR products (Fig. 1b). The transposition of mini-Tn7 was orientation-specific with Tn7R located immediately downstream of and facing glmS1 (Fig. 1a). Site- and orientation-specific transposition is a common feature of Tn7 insertions into other bacterial chromosomes (Choi & Schweizer, 2006; Choi & Schweizer, 2006; Choi et al., 2006; Choi et al., 2005; Choi et al., 2008; Waddell & Craig, 1989). Possible insertions of mini-Tn7 at the glmS2 site were assessed using PCR amplification with glmS2Up (5’GATGGCGACCTGCCGCTG3’) and glmS2Down (5’GCATCGTCGGCCGCGACA3’) (Fig. 1a). Amplification from all strains resulted in a 283 bp PCR product that indicates that no insertion of mini-Tn7 occurred at the glmS2 site (Fig. 1b). Based on the DNA sequence analysis of the PCR products from five reactions, the insertion site of mini-Tn7 was located 25 nucleotides downstream of the glmS1 stop codon (Fig. 1c). The analysis of the 3’ glmS1 nucleotide sequence identified a putative attTn7 site that was very similar to the attTn7 sites identified in E. coli (Waddell & Craig, 1989) and P. aeruginosa (Choi & Schweizer, 2006) (Fig. 1c). No putative attTn7 site could be identified at glmS2 (Fig. 1c). These results support the experimental findings that mini-Tn7 insertions seemed to be confined to the glmS1-linked attTn7 site with no insertions observed at glmS2.
Fig. 1. Transposition of mini-Tn7 into the X. campestris pv. campestris (Xcc) chromosome.
(A) Physical maps of Xcc glmS1 and glmS2 loci (top and bottom) and an integration event at glmS1 (Xcc::mini-Tn7T-Gm; middle). Arrows denote the positions and orientations of the indicated oligonucleotide primers. Tn7R and Tn7L, right and left end of Tn7, respectively; FRT, FLP recombinase target; aacC1, Gm acetyltransferase-encoding gene.
(B) PCR amplification to confirm the transposition of the mini-Tn7T-Gm. Genomic DNA of a representative Gmr transformant was used as the template in PCR reactions with primer pairs specific for glmS1 and glmS2 as follows: lane 1, glmS1Up and Tn7R; lane 2, glmS1Down and Tn7L; lane 3, glmS1Up and Tn7L; lane 4, glmS1Down and Tn7R; lane 5, glmS2Up and Tn7R; lane 6, glmS2Down and Tn7L; lane 7, glmS2Up and Tn7L; lane 8, glmS2Down and Tn7R; and lane 9, glmS2Up and glmS2Down. M1 and M2 are 100-bp molecular weight ladder (Fermentas, Canada) for lanes 1 to 4, and lanes 5 to 9, respectively.
(C) Alignment of the nucleotide sequences of the glmS 3’ and downstream sequences. The sequences analyzed were the glmS regions of E. coli (Waddell & Craig, 1989) (Eco) and P. aeruginosa (Choi & Schweizer, 2006) (Pae), and the putative glmS1 and glmS2 regions of Xcc [xcc0569 and xcc3411 (da Silva et al., 2002)], Xoo [xoo0746 and xoo3917 (Lee et al., 2005)], X. campestris pv. vesicatoria [xcv3754 and xcv0770 (Thieme et al., 2005)] and X. axonopodis pv. citri [xac3637 and xac0714 (da Silva et al., 2002)]. attTn7 associated sequences (boxed) were identified based on a previous study with E. coli (Waddell & Craig, 1989) and asterisks represent sequence identity in all species. Arrowheads indicate mini-Tn7 insertion sites. Bold and gray letters denote the stop codons of the respective glmS genes.
The excision of the Gmr marker
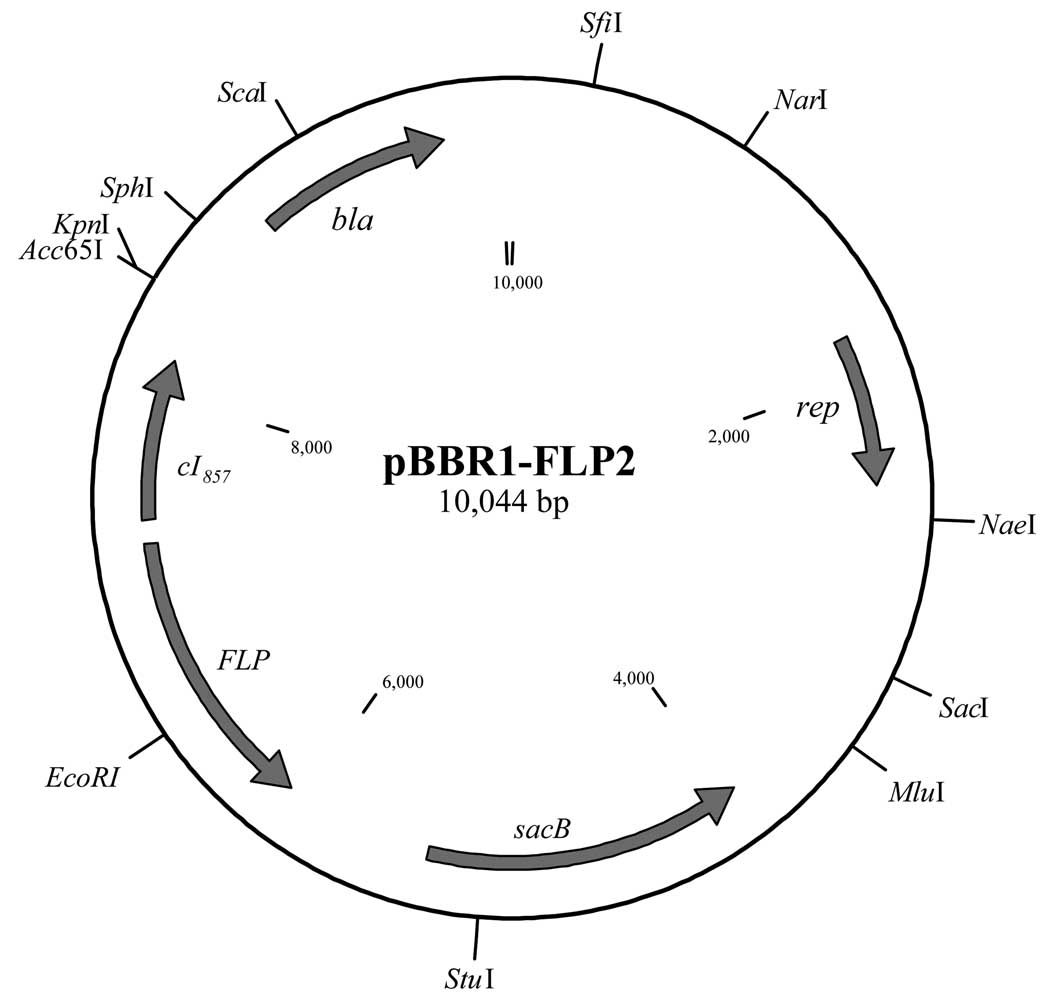
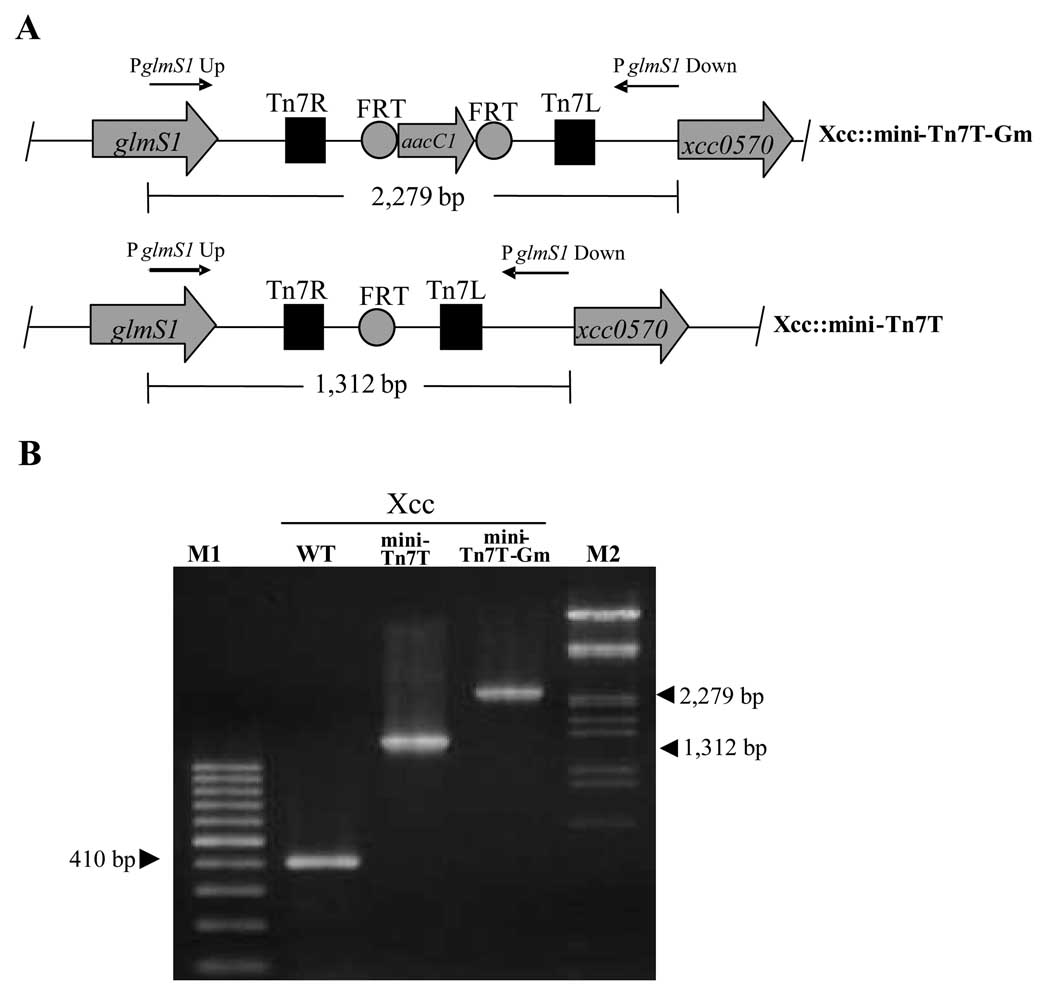
pUC18-mini-Tn7T-Gm, a mini-Tn7 vector, contains two FLP recombinase target (FRT) sites flanking aacC1 (Gmr) (Fig. 1a). These FRT sites allow for flipase (FLP) mediated excision of aacC1. To assess whether an unmarked mini-Tn7 insertion could be generated in Xcc, pFLP2 (Hoang et al., 1998) was introduced into three representative Xcc transformants (Xcc::mini-Tn7T-Gm). The pFLP2 plasmid encodes the Saccharomyces cerevisiae FLP recombinase enzyme whose expression is driven by a λ promoter under the control of temperature sensitive λ repressor (cI857). After several attempts to transform pFLP2 into Xcc::mini-Tn7T-Gm, no transformants could be obtained, suggesting that the plasmid may not be able to efficiently replicate and be maintained in Xcc. Hence, a cI857-FLP-sacB containing fragment of pFLP2 was sub-cloned on a Acc65I-SphI (blunt) fragment into the broad-host range pBBR1MCS-4 plasmid which contains a carbenicillin resistance (Cbr) gene. The resulting expression vector pBBR1-FLP2 (Fig. 2) is capable of expressing cloned genes and replicating in Xanthomonas, yielding pBBR1-FLP2. This plasmid was subsequently electroporated into Xcc::mini-Tn7T-Gm and Cbr transformants were selected. Excision of the Gm resistance cassette resulted in Cbr and Gms transformants, and the excision event was confirmed by PCR analysis. PCR analyses using Xcc::mini-Tn7T-Gm genomic DNA and primers glmS1Up and glmS1Down generated a PCR product of 2,270 bp (Fig. 3a and b). Excision of aacC1 by FLP-mediated recombination should result in a PCR product of 1,312 bp when amplification is performed using glmS1Up and glmS1Down primers and DNA from Cbr and Gms colonies. As expected, 1,312 bp PCR products were detected (Fig. 3b). Hence, pBBR1-FLP2 could be efficiently used to mediate FLP-FRT recombination in order to generate an unmarked Tn7 insertion in Xanthomonas. Moreover, the advantage of using pBBR1MCS and its derivative plasmids is that they can stably replicate at a moderate copy number in a number of Gram-negative bacteria, including α- and γ-proteobacteria (Khan et al., 2008; Kovach et al., 1995; Vattanaviboon et al., 2007). Also, these plasmids can be mobilized through the IncP1 conjugative transfer system into many Gram-negative bacteria (Kovach et al., 1995). The removal pBBR1-FLP2 plasmid was cured from Xcc::mini-Tn7T was by growing the strain on SB medium containing 5% sucrose. pBBR1-FLP2 contains a counter-selection gene, sacB, which encodes Bacillus subtilis levansucrase (Gay et al., 1985). The presence of sucrose in the medium causes cell death in Gram-negative bacteria due to production of a toxic high-molecular-weight fructose polymer. Cells which survived and grew on the sucrose-supplemented medium were selected for further characterization and were shown to be Cbs indicating plasmid loss. The pBBR1-FLP2-cured clones could be obtained after a single passage of cells through SB broth supplemented with sucrose. Therefore, the presence of the sacB counter-selection marker in pBBR1-FLP2 allows for the simple and rapid elimination of the plasmid from the Xcc host.
Fig. 2. Map of FLP recombinase expressing plasmid.
The physical map of pBBR1-FLP2 (GenBank accession number FJ797950) shows the locations and orientations of the genetic determinants involved in replication (rep), mobilization (mob), ampicillin/ carbenicillin resistance (bla, β-lactamase-coding gene), FLP-FRT recombination (FLP, Flippase-coding gene) and the counter-selectable marker (sacB, Bacillus subtilis levansucrase-encoding gene). Some unique restriction enzyme sites are shown. cI857, gene encoding a temperature sensitive λ repressor.
Fig. 3. FLP-mediated excision of antibiotic resistance gene.
(A) Physical maps of Xcc::mini-Tn7T-Gm and Xcc::mini-Tn7T showing the insertion at the glmS1 locus. Arrows represent the positions and orientations of oligonucleotide primers used in PCR analysis of excision events. Tn7R and Tn7L, right and left end of Tn7, respectively; FRT, FLP recombinase target; aacC1, Gm acetyltransferase-encoding gene.
(B) PCR amplification to confirm the excision of the Gmr marker. Genomic DNAs of Xcc::mini-Tn7T-Gm and Xcc::mini-Tn7T were amplified with glmS1Up and glmS1Down primers. Arrowheads show the migration of expected DNA fragments whose sizes are indicated in kilobases. M1 and M2 are a 100 bp DNA ladder and the λDNA/EcoRI+HindIII markers (Fermentas, Canada), respectively. WT, Xcc wild-type.
Insertion of mini-Tn7 elements did not affect Xcc virulence
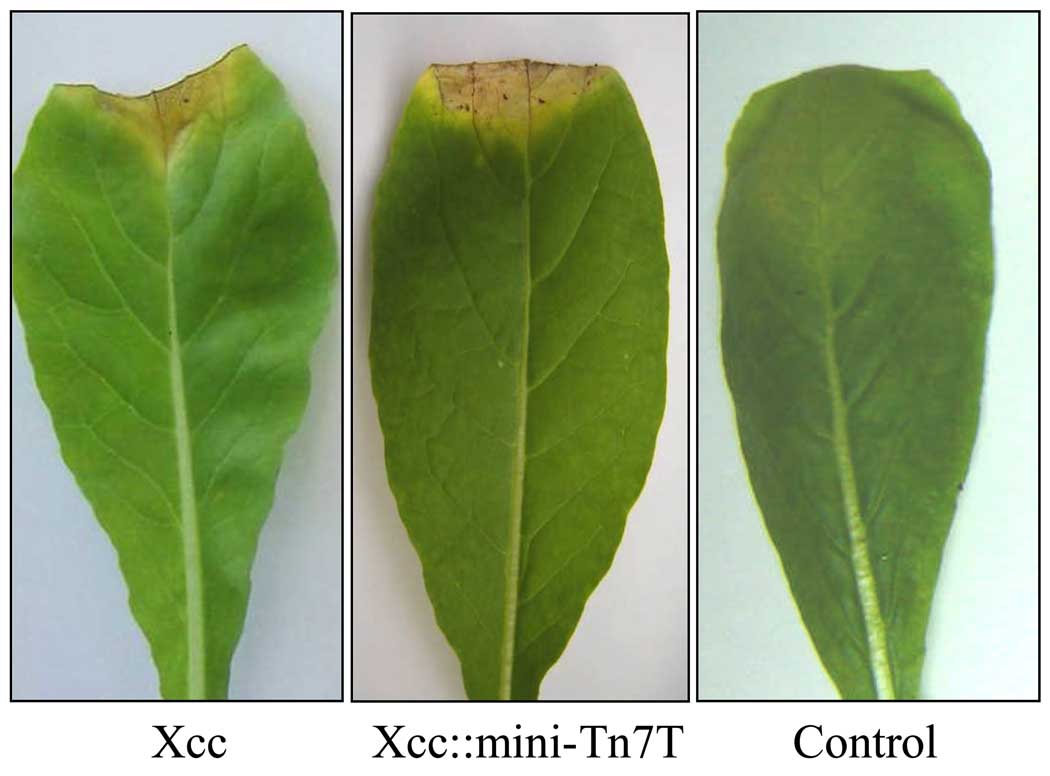
Xcc is a causative agent of black rot disease in cruciferous crops. The usefulness of the mini-Tn7 system as a genetic tool in Xanthomonas and in the investigation of plant--microbe interactions depends on the premise that mini-Tn7 insertion should not affect the virulence of the bacteria on the compatible host plant. Wild-type Xcc and Xcc::mini-Tn7T strains were inoculated into Chinese radish (Raphanus sativus) leaves by the leaf clipping method (Dow et al., 2003). The results demonstrate that the lesion length in both Xcc::mini-Tn7T and the isogenic wild-type strains was not significantly different (P-value > 0.05). This suggests that the transposition of the mini-Tn7 into the glmS1 site has no effect on the virulence of Xcc on the tested host plant (Fig. 4). Thus, the mini-Tn7 offers a potential shuttle vector system for the site-specific chromosomal integration for single copy complementation experiments, as well as gene expression analysis in X. campestris pv. campestris using reporter gene constructs.
Fig. 4. Virulence testing in X. campestris pv. campestris strains.
The virulence of Xcc and Xcc::mini-Tn7T strains was tested on a susceptible host, Chinese radish (Raphanus sativus) (Dow et al., 2003). The infection of the tested plant was performed using the leaf clipping method (Dow et al., 2003). Lesion lengths were measured 14 days post-inoculation. Control is the leaf without bacterial inoculation.
Gene expression analysis in Xcc using mini-Tn7 vector
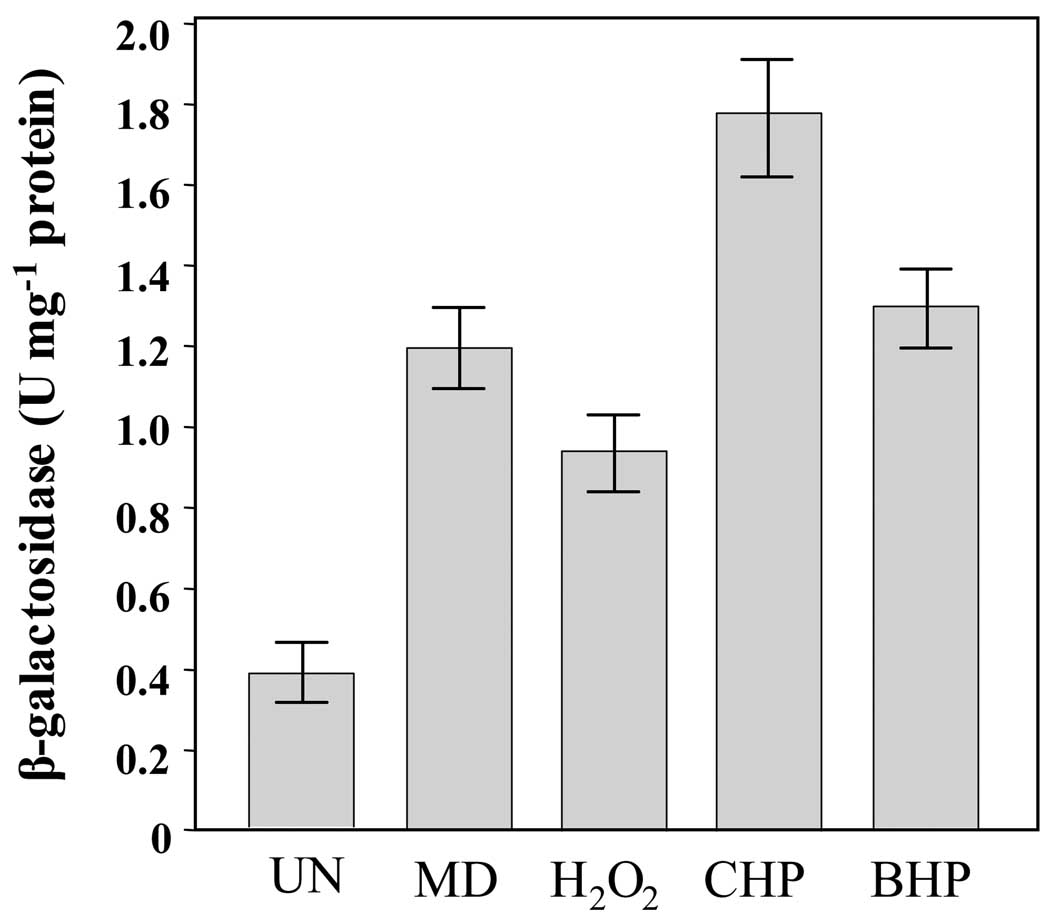
We have characterized ahpC, a gene encoding alkyl hydroperoxide reductase, in X. campestris pv. phaseoli (Loprasert et al., 2000; Mongkolsuk et al., 1997). The expression of ahpC is inducible by H2O2, organic hydroperoxides and superoxide generators (menadione and paraquat) in an OxyR dependent manner. In an attempt to evaluate the use of mini-Tn7 vector in gene expression analysis in Xcc, ahpC promoter was transcriptionally fused to lacZ in pUC18-mini-Tn7T-Gm-lacZ as described in the Materials and Methods. The recombinant plasmid pTn7T-PahpC-lacZ was introduced into Xcc wild-type and the transformants were selected for Gmr phenotype. Insertion of Tn7 elements containing ahpC promoter-lacZ fusion and curing of pBBR1-FLP2 from the transformants giving Xcc::mini-Tn7T-PahpC-lacZ, was verified as described in an earlier section. The ahpC promoter activity was monitored by measuring β-galactosidase activity. Xcc::mini-Tn7T-PahpC-lacZ was cultivated in SB medium until the cells reached exponential phase of growth before bacterial cultures were being challenged 100 µM menadione (MD), 100 µM H2O2, 100 µM cumene hydroperoxide (CHP) 100 µM tert-butyl hydroperoxide (BHP). As illustrated in Fig. 5, treatment with MD, H2O2, CHP and BHP increased β-galactosidase activity by 3.0, 2.4, 4.5 and 3.3 folds, respectively, relative to wild-type level. The expression pattern of ahpC in Xcc was similar to previously characterized and closely related ahpC from X. campestris pv. phaseoli (Mongkolsuk et al., 1997). Our data presented the usefulness of mini-Tn7 vectors as tools to assess gene expression profile in plant pathogenic Xanthomonas.
Fig. 5. Expression analysis of ahpC in X. campestris pv. campestris.
Xcc::mini-Tn7T-PahpC-lacZ (the ahpC promoter transcriptionally fused to lacZ reporter inserted into chromosome on a mini-Tn7 vector) cultures were grown either uninduced or induced with 100 µM of menadione (MD), H2O2, cumene hydroperoxide (CHP) or tert-butyl hydroperoxide (BHP) for 30 min. The β-galactosidase activity in clear lysates was determined and expressed as international units per mg protein. UN, un-induced.
Application of the mini-Tn7 vectors in other Xanthomonas
We further tested the functionality of the mini-Tn7 system in other Xanthomonas spp., specifically Xanthomonas oryzae pv. orzyae (Xoo). The pUC18-mini-Tn7T-Gm delivery plasmid and pTNS2 helper plasmid were introduced into Xoo by electroporation using previously described conditions (Mongkolsuk et al., 1996). The mini-Tn7 insertion sites were determined in 35 transformants by PCR using genomic DNA templates isolated and two pairs of primers (glmS1XoUp, 5’-CGACCGCCTGCTGGAAAA3’, and Tn7R or glmS1Xo Down, 5’-ATCTTCGACGCTCAACAG3’, and Tn7L). Amplification of a 280 bp fragment demonstrated that insertion of the mini-Tn7T-Gm element in all transformants occurred downstream of glmS1 (xoo0746). Furthermore, sequence analysis of the PCR products indicated that the mini-Tn7 insertion site (attTn7) was 25 bp downstream of glmS1. No insertions were observed at glmS2 (xoo3917)(data not shown). We also analyzed the published genomic sequences of all Xanthomonas spp. and determined that X. campestris pv. vesicatoria (Thieme et al., 2005), X. oryzae pv. oryzae (Xoo) (Lee et al., 2005), and X. axonopodis pv. citri (da Silva et al., 2002) all contain two putative glmS genes. However, upon searching for putative attTn7 sites surrounding the two glmS genes, attTn7 sites could only be identified downstream of the respective glmS1 genes (Fig. 1c). Based on the results obtained with the mini-Tn7 system in Xcc and Xoo, it is likely that the mini-Tn7 system will work in other Xanthomonas spp. and that the mini-Tn7 will probably insert downstream of glmS1. Additionally, the excision of aacC1 from Xoo::mini-Tn7T-Gm was tested by transforming the strain with pBBR1-FLP2. The results from the PCR analysis using glmS1XoUp and glmS1XoDown primers and DNA from Xoo::mini-Tn7T/pBBR1-FLP2 demonstrated the excision of the Gmr marker through FLP-FRT recombination (data not shown). As described in the earlier section 3.2, pBBR1-FLP2 could be cured from the resulting strain with sucrose counter-selection. Taken together, our data suggest the possible applications of mini-Tn7 system and a new vector for FLP-FRT system for making unmarked mutation in a variety species of xanthomonads. This will be useful for genetic analysis of industrially important and phytopathogenic bacteria, Xanthomonas spp.
Acknowledgements
The research was supported by grants (BT-B-01-PG-14-5112) from the National Center for Genetic Engineering and Biotechnology (BIOTEC), Mahidol University and from the ESTM under the higher Education Development Project of the Ministry of Education. T.J. was supported by a Royal Golden Jubilee Scholarship PHD/0222/2547 from the Thailand Research Fund. HPS was supported by NIH grants AI058141 and AI065357.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arciszewska LK, Drake D, Craig NL. Transposon Tn7: cis-Acting sequences in transposition and transposition immunity. J Mol Biol. 1989;207:35–52. doi: 10.1016/0022-2836(89)90439-7. [DOI] [PubMed] [Google Scholar]
- Chauvatcharin N, Atichartpongkul S, Utamapongchai S, Whangsuk W, Vattanaviboon P, Mongkolsuk S. Genetic and physiological analysis of the major OxyR-regulated katA from Xanthomonas campestris pv. phaseoli. Microbiology. 2005;151:597–605. doi: 10.1099/mic.0.27598-0. [DOI] [PubMed] [Google Scholar]
- Chilton MD, Currier TC, Farrand SK, Bendich AJ, Gordon MP, Nester EW. Agrobacterium tumefaciens DNA and PS8 bacteriophage DNA not detected in crown gall tumors. Proc Natl Acad Sci USA. 1974;71:3672–3676. doi: 10.1073/pnas.71.9.3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KH, Schweizer HP. Mini-Tn7 insertion in bacteria with secondary, non-glmS-linked attTn7 sites: example Proteus mirabilis HI4320. Nat Protoc. 2006;1:170–178. doi: 10.1038/nprot.2006.26. [DOI] [PubMed] [Google Scholar]
- Choi KH, Schweizer HP. Mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat Protoc. 2006;1:153–161. doi: 10.1038/nprot.2006.24. [DOI] [PubMed] [Google Scholar]
- Choi KH, DeShazer D, Schweizer HP. Mini-Tn7 insertion in bacteria with multiple glmS-linked attTn7 sites: example Burkholderia mallei ATCC 23344. Nat Protoc. 2006;1:162–169. doi: 10.1038/nprot.2006.25. [DOI] [PubMed] [Google Scholar]
- Choi KH, Gaynor JB, White KG, Lopez C, Bosio CM, Karkhoff-Schweizer RR, Schweizer HP. A Tn7-based broad-range bacterial cloning and expression system. Nat Methods. 2005;2:443–448. doi: 10.1038/nmeth765. [DOI] [PubMed] [Google Scholar]
- Choi KH, Mima T, Casart Y, Rholl D, Kumar A, Beacham IR, Schweizer HP. Genetic tools for select-agent-compliant manipulation of Burkholderia pseudomallei. Appl Environ Microbiol. 2008;74:1064–1075. doi: 10.1128/AEM.02430-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva AC, Ferro JA, Reinach FC, Farah CS, Furlan LR, Quaggio RB, Monteiro-Vitorello CB, Van Sluys MA, Almeida NF, Alves LM, do Amaral AM, Bertolini MC, Camargo LE, Camarotte G, Cannavan F, Cardozo J, Chambergo F, Ciapina LP, Cicarelli RM, Coutinho LL, Cursino-Santos JR, El-Dorry H, Faria JB, Ferreira AJ, Ferreira RC, Ferro MI, Formighieri EF, Franco MC, Greggio CC, Gruber A, Katsuyama AM, Kishi LT, Leite RP, Lemos EG, Lemos MV, Locali EC, Machado MA, Madeira AM, Martinez-Rossi NM, Martins EC, Meidanis J, Menck CF, Miyaki CY, Moon DH, Moreira LM, Novo MT, Okura VK, Oliveira MC, Oliveira VR, Pereira HA, Rossi A, Sena JA, Silva C, de Souza RF, Spinola LA, Takita MA, Tamura RE, Teixeira EC, Tezza RI, Trindade dos Santos M, Truffi D, Tsai SM, White FF, Setubal JC, Kitajima JP. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature. 2002;417:459–463. doi: 10.1038/417459a. [DOI] [PubMed] [Google Scholar]
- Dow JM, Crossman L, Findlay K, He YQ, Feng JX, Tang JL. Biofilm dispersal in Xanthomonas campestris is controlled by cell-cell signaling and is required for full virulence to plants. Proc Natl Acad Sci USA. 2003;100:10995–11000. doi: 10.1073/pnas.1833360100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay P, Le Coq D, Steinmetz M, Berkelman T, Kado CI. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J Bacteriol. 1985;164:918–921. doi: 10.1128/jb.164.2.918-921.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212:77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- Khan SR, Gaines J, Roop RM, 2nd, Farrand SK. Broad-host-range expression vectors with tightly regulated promoters and their use to examine the influence of TraR and TraM expression on Ti plasmid quorum sensing. Appl Environ Microbiol. 2008;74:5053–5062. doi: 10.1128/AEM.01098-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, 2nd, Peterson KM. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- Lee BM, Park YJ, Park DS, Kang HW, Kim JG, Song ES, Park IC, Yoon UH, Hahn JH, Koo BS, Lee GB, Kim H, Park HS, Yoon KO, Kim JH, Jung CH, Koh NH, Seo JS, Go SJ. The genome sequence of Xanthomonas oryzae pathovar oryzae KACC10331, the bacterial blight pathogen of rice. Nucleic Acids Res. 2005;33:577–586. doi: 10.1093/nar/gki206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loprasert S, Fuangthong M, Whangsuk W, Atichartpongkul S, Mongkolsuk S. Molecular and physiological analysis of an OxyR-regulated ahpC promoter in Xanthomonas campestris pv. phaseoli. Mol Microbiol. 2000;37:1504–1514. doi: 10.1046/j.1365-2958.2000.02107.x. [DOI] [PubMed] [Google Scholar]
- Mongkolsuk S, Loprasert S, Whangsuk W, Fuangthong M, Atichartpongkun S. Characterization of transcription organization and analysis of unique expression patterns of an alkyl hydroperoxide reductase C gene (ahpC) and the peroxide regulator operon ahpF-oxyR-orfX from Xanthomonas campestris pv. phaseoli. J Bacteriol. 1997;179:3950–3955. doi: 10.1128/jb.179.12.3950-3955.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongkolsuk S, Loprasert S, Vattanaviboon P, Chanvanichayachai C, Chamnongpol S, Supsamran N. Heterologous growth phase- and temperature-dependent expression and H2O2 toxicity protection of a superoxide-inducible monofunctional catalase gene from Xanthomonas oryzae pv. oryzae. J Bacteriol. 1996;178:3578–3584. doi: 10.1128/jb.178.12.3578-3584.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panmanee W, Vattanaviboon P, Eiamphungporn W, Whangsuk W, Sallabhan R, Mongkolsuk S. OhrR, a transcription repressor that senses and responds to changes in organic peroxide levels in Xanthomonas campestris pv. phaseoli. Mol Microbiol. 2002;45:1647–1654. doi: 10.1046/j.1365-2958.2002.03116.x. [DOI] [PubMed] [Google Scholar]
- Peters JE, Craig NL. Tn7: smarter than we thought. Nat Rev Mol Cell Biol. 2001;2:806–814. doi: 10.1038/35099006. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Thieme F, Koebnik R, Bekel T, Berger C, Boch J, Buttner D, Caldana C, Gaigalat L, Goesmann A, Kay S, Kirchner O, Lanz C, Linke B, McHardy AC, Meyer F, Mittenhuber G, Nies DH, Niesbach-Klosgen U, Patschkowski T, Ruckert C, Rupp O, Schneiker S, Schuster SC, Vorholter FJ, Weber E, Puhler A, Bonas U, Bartels D, Kaiser O. Insights into genome plasticity and pathogenicity of the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria revealed by the complete genome sequence. J Bacteriol. 2005;187:7254–7266. doi: 10.1128/JB.187.21.7254-7266.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vattanaviboon P, Tanboon W, Mongkolsuk S. Physiological and expression analyses of Agrobacterium tumefaciens trxA, encoding thioredoxin. J Bacteriol. 2007;189:6477–6481. doi: 10.1128/JB.00623-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell CS, Craig NL. Tn7 transposition: recognition of the attTn7 target sequence. Proc Natl Acad Sci USA. 1989;86:3958–3962. doi: 10.1073/pnas.86.11.3958. [DOI] [PMC free article] [PubMed] [Google Scholar]