Figure 2.
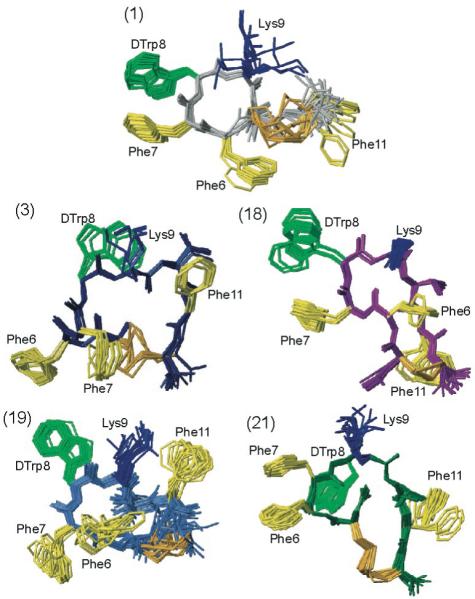
The NMR structures of the five analogues studied by NMR (i.e., analogues 1, 3, 18, 19 and 21 as indicated). For each analogue, twenty energy-minimized conformers with the lowest target function are used to represent the 3D NMR structure. The bundle is obtained by overlapping the Cα atoms of all the residues. The backbone and the side chains are displayed including the disulfide bridge. The following color code is used: grey (1) H-c[Cys-Phe-Phe-dTrp-Lys-Thr-Phe-Cys]-OH, ODT-8 taken from Grace et al.;34 navy-blue (3)H-c[Cys-Phe-Phe-dTrp-Lys-Thr-Phe-dCys]-OH; violet (18) Ac-c[dNcy-Phe-Phe-dTrp-Lys-Thr-Phe-Ncy]-OH; royal blue (19) Ac-c[Ncy-Phe-Phe-dTrp-Lys-Thr-Phe-Ncy]-OH; dark green (21) Ac-c[Hcy-Phe-Phe-dTrp-Lys-Thr-Phe-Cys]-OH. The amino acid side chains which are proposed to be involved in binding to the various SRIF receptors are highlighted: dTrp at position 8 in light green, Lys at position 9 in blue, and Phe at positions 6, 7 and 11 yellow. The disulphide bridges are shown in orange for clarity.
