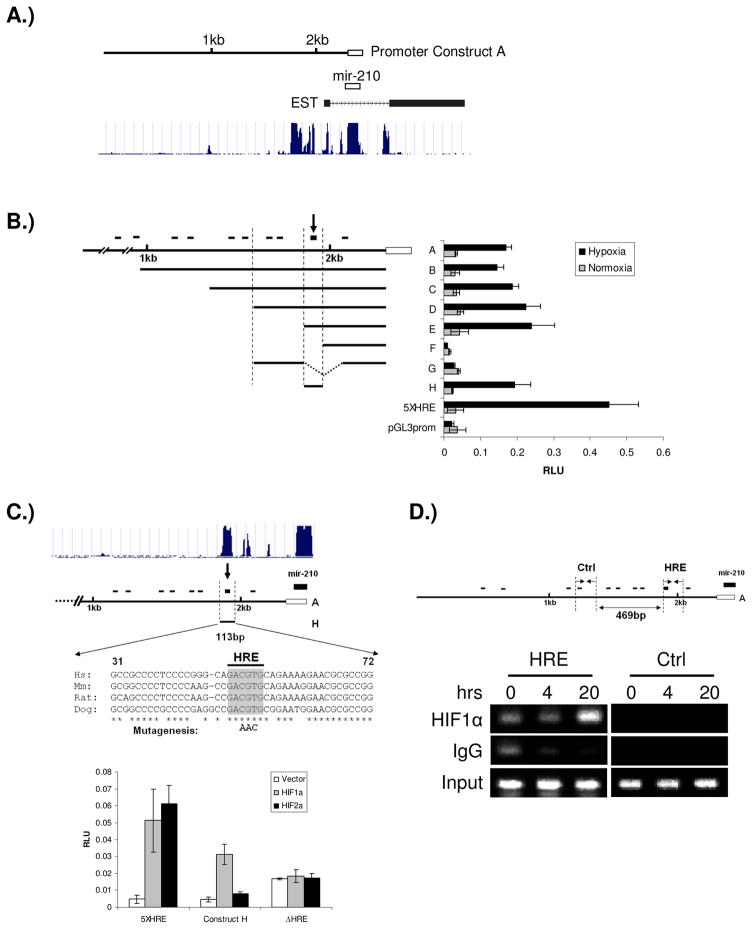
Figure 3.
Identification of the functional HRE in mir-210 promoter. A) The schematic view of mir-210 genomic structure. The location of the cloned promoter related to mir-210 coding region was shown in top panel. Open box indicates the coding region of mir-210 and solid line indicates the 2.3kb promoter cloned. The conservation of mir-210 genomic sequences across species was shown in the bottom panel, which is from the UCSC database (http://genome.ucsc.edu). B) mir-210 promoter analysis. Promoter fragments were cloned into the pGL3-Basic vector; a 5XHRE construct, including five copies of the human VEGF HRE, serves as a positive control. Serial deletion constructs (identified by the letters A–H in the middle of the figure) are schematically represented in the bar diagram on the left, while their relative promoter activities in normoxia (gray bars) and 2% oxygen (black bars) are displayed on the right of the figure. The small black bars on the top represent the relative location of the ten potential HREs. The arrow at the top of the bar diagram indicates this HRE is essential for up-regulation of promoter activity under hypoxia. The open box represents mir-210 coding sequence; C) Mutation of the HRE sequence abolishes mir-210 promoter’s responsiveness to HIF1α. Constructs A and H are shown. Upper panel shows that the sequence surrounding the HRE site is the only region highly conserved across species in mir-210 promoter according to the UCSC database. The conserved nucleotides from 31–72 in construct H are indicated by asterisks. Three nucleotide mutations in the core of the HRE are in bold letters. Luciferase assays of construct H with/without HRE mutation are shown in the lower panel. Error bars indicate standard deviation. D) ChIP analysis of the mir-210 promoter. Upper panel, the relative locations of ChIP primers on mir-210 promoter. Lower panel, ChIP results. HSC3 cells were cultured under normoxia or 0.5% oxygen for 4 and 20 hours, respectively. PCR was performed with primers specific to the functional mir-210 HRE (HRE) and a region that is ~500bp upstream of the HRE site serving as a control (Ctrl). IgG refers to samples derived for the IP-negative control, HIF1α refers to DNA immunoprecipitated with a HIF1α antibody, and Input refers to the DNA derived from samples prior to immunoprecipitation.