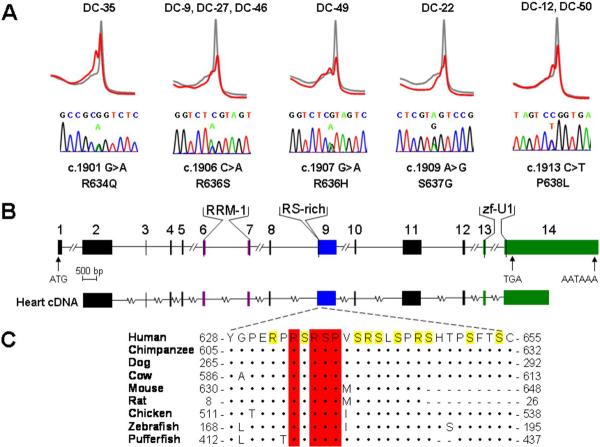
Figure 3. RBM20 Mutations Identified in Families with Dilated Cardiomyopathy.
(A) Genomic DNA mutation scans using DHPLC revealed abnormal heteroduplex profiles (red), compared to the control wild-type profile (gray), in exon 9 of RBM20 in 8 DCM families. Abnormal profiles indicated DNA sequence alterations, which were absent in 480 control samples. Below the heteroduplex profiles, DNA sequencing revealed corresponding heterozygous missense mutations. The location of each mutation, and its resultant amino acid substitution, are based on predicted reference RBM20 cDNA (mRNA) and protein sequences and are indicated below each chromatogram. Mutation c.1906 C>A, R636S was shared by three families and c.1913 C>T, P638L was shared by two families. (B) The predicted genomic structure of RBM20, consisting of 14 exons, is depicted to scale. Exons that encode peptides homologous to highly conserved functional domains – RNA-recognition motif 1 (RRM-1, in purple), arginine/serine-rich region (RS-rich, in blue), and U1 zinc finger (zf-U1, in green) – are indicated. Putative start (ATG) and stop (TGA) codons are located in exons 1 and 14, respectively. A polyadenylation signal (AATAAA) is located at the 3' end of exon 14. Directly below the RBM20 genomic structure, cDNA amplification and sequencing confirmed transcription of mRNA from exons 2–14 in human heart tissue, as depicted by parallel alignment. The cDNA transcript contains the complete RS domain and identified RBM20 mutations in the 5' region of exon 9. (C) Alignment of homologous RBM20 protein sequences that flank the amino acid substitutions is shown. The RS domain spans residues 632–654, with arginine (R) and serine (S) residues highlighted in yellow. Residues conserved between human RBM20 and another species are indicated by (•) and amino acid deletions by (-). Amino acids that are altered by the identified RBM20 missense mutations (residues 634, 636, 637, and 638, indicated with red bars) are conserved among all eight species. Accession numbers: XP_291671 for human, XP_508032 for chimpanzee, XP_544017 for dog, XP_603772 for cow, BAE24961 for mouse, NP_001101081 for rat, XP_421755 for chicken, XP_683222 for zebrafish, and CAG01297 for pufferfish.